7PEG
 
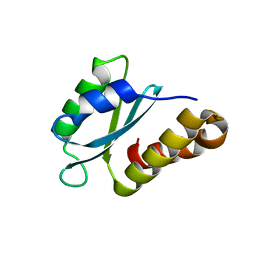 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZY
 
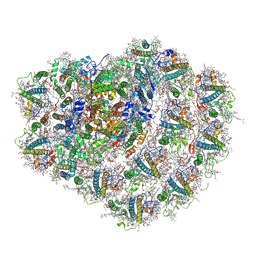 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZX
 
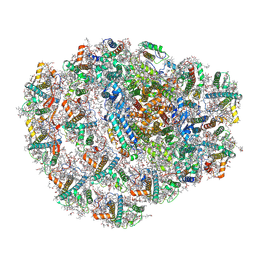 | | Structure of low-light grown Chlorella ohadii Photosystem I | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7PLM
 
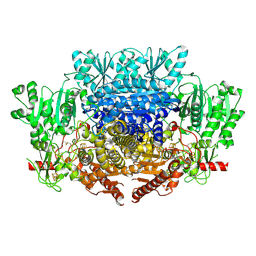 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
8CK0
 
 | |
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
6HIN
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, F conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIQ
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIO
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIS
 
 | | Mouse serotonin 5-HT3 receptor, tropisetron-bound, T conformation | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
