8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
3J7H
 
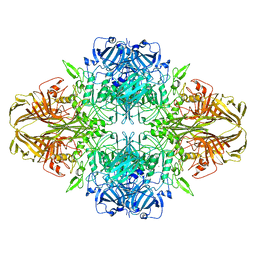 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | 分子名称: | Beta-galactosidase, MAGNESIUM ION | | 著者 | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JD2
 
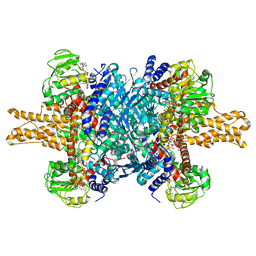 | | Glutamate dehydrogenase in complex with NADH, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
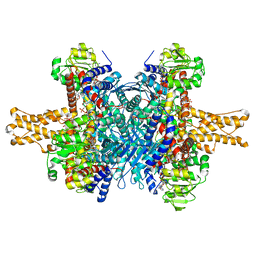 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
4YJ0
 
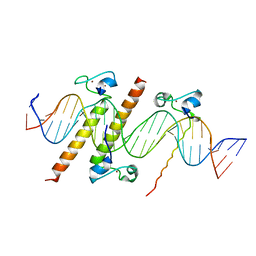 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | 分子名称: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | 著者 | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | 登録日 | 2015-03-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (3.814 Å) | | 主引用文献 | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5A6Q
 
 | | Native structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, ... | | 著者 | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | 登録日 | 2015-06-30 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
5A6Z
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with lewis a | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, LECB, ... | | 著者 | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | 登録日 | 2015-07-02 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
7NTL
 
 | | AA9 lytic polysaccharide monooxygenase (LPMO) from Malbranchea cinnamomea | | 分子名称: | CITRIC ACID, COPPER (II) ION, LPMO9F | | 著者 | Mazurkewich, S, Seveso, A, Huttner, S, Branden, G, Larsbrink, L. | | 登録日 | 2021-03-10 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure of a C1/C4-oxidizing AA9 lytic polysaccharide monooxygenase from the thermophilic fungus Malbranchea cinnamomea.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8P7I
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7N
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | FORMIC ACID, Parathion hydrolase, ZINC ION | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7V
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | 著者 | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.737 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7Q
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7R
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7S
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7M
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | FORMIC ACID, GLYCEROL, METHYLPHOSPHONIC ACID ESTER GROUP, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7U
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7H
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1,2-ETHANEDIOL, 2-methylidene-1,3-dioxane-4,4-diol, FORMIC ACID, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.774 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7T
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7F
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | Parathion hydrolase, ZINC ION | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4Z0L
 
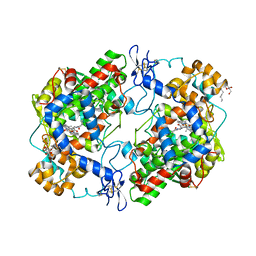 | | The murine cyclooxygenase-2 complexed with a nido-dicarbaborate-containing indomethacin derivative | | 分子名称: | (R)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, (S)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, S, Neumann, W, Banerjee, S, Hey-Hawkins, E, Marnett, L.J. | | 登録日 | 2015-03-26 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | nido-Dicarbaborate Induces Potent and Selective Inhibition of Cyclooxygenase-2.
Chemmedchem, 11, 2016
|
|
8P7K
 
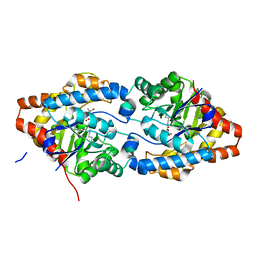 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | (2~{R})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | 著者 | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.929 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5A1A
 
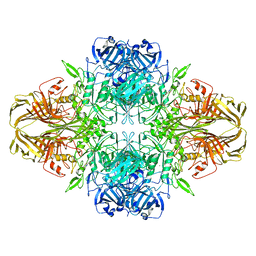 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | 著者 | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | 登録日 | 2015-04-29 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
6E5L
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiol (abn-CBD) | | 分子名称: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | 著者 | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-20 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
