2TMV
 
 | |
6LY8
 
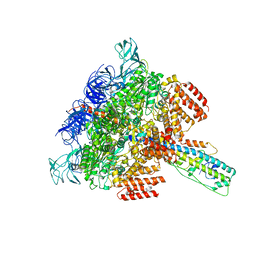 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
3TEE
 
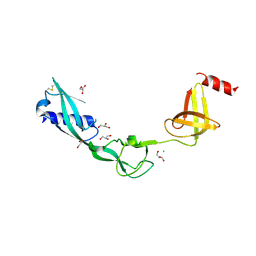 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
6LY9
 
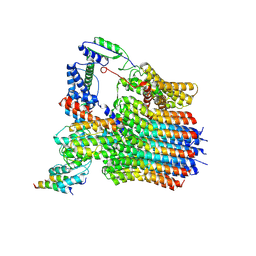 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
3VJP
 
 | |
3VKI
 
 | |
3AJW
 
 | | Structure of FliJ, a soluble component of flagellar type III export apparatus | | Descriptor: | Flagellar fliJ protein, MERCURY (II) ION | | Authors: | Imada, K, Ibuki, T, Minamino, T, Namba, K. | | Deposit date: | 2010-06-23 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases
Nat.Struct.Mol.Biol., 18, 2011
|
|
6JY0
 
 | | CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V) | | Descriptor: | Flagellin | | Authors: | Yamaguchi, T, Toma, S, Terahara, N, Miyata, T, Minamino, T, Ashikara, M, Namba, K, Kato, T. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural and Functional Comparison ofSalmonellaFlagellar Filaments Composed of FljB and FliC.
Biomolecules, 10, 2020
|
|
3AJC
 
 | | Structure of the MC domain of FliG (PEV), a CW-biased mutant | | Descriptor: | Flagellar motor switch protein fliG | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rotational switching mechanism of the bacterial flagellar motor
Plos Biol., 9, 2011
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7EHA
 
 | | Crystal structure of the flagellar hook cap from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
7EH9
 
 | | Crystal structure of the flagellar hook cap fragment from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
5WRH
 
 | |
7W2J
 
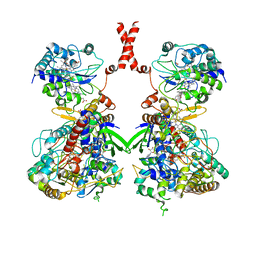 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
5XL0
 
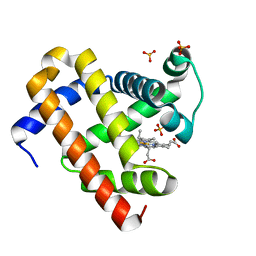 | | met-aquo form of sperm whale myoglobin reconstituted with 7-PF, a heme possesseing CF3 group as side chain | | Descriptor: | Myoglobin, SULFATE ION, fluorinated heme | | Authors: | Kanai, Y, Harada, A, Shibata, T, Nishimura, R, Namiki, K, Watanabe, M, Nakamura, S, Yumoto, F, Senda, T, Suzuki, A, Neya, S, Yamamoto, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of Heme Orientational Disorder in a Myoglobin Reconstituted with a Trifluoromethyl-Group-Substituted Heme Cofactor
Biochemistry, 56, 2017
|
|
1WLG
 
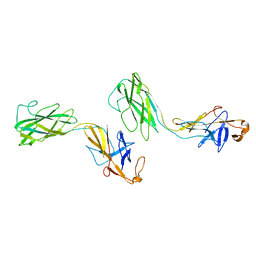 | | Crystal structure of FlgE31, a major fragment of the hook protein | | Descriptor: | Flagellar hook protein flgE | | Authors: | Samatey, F.A, Matsunami, H, Imada, K, Nagashima, S, Shaikh, T.R, Thomas, D.R, DeRosier, D.J, Kitao, A, Namba, K. | | Deposit date: | 2004-06-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism.
Nature, 431, 2004
|
|
2SDF
 
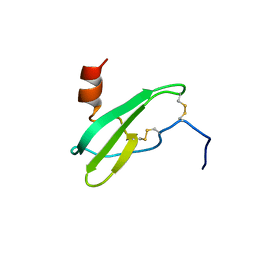 | | SOLUTION NMR STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), 30 STRUCTURES | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-03-07 | | Release date: | 1998-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
7WSQ
 
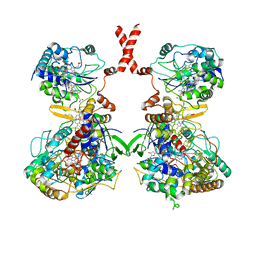 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
3RI6
 
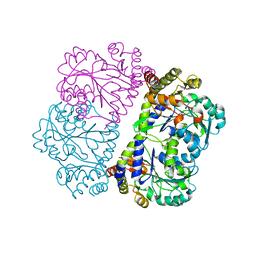 | | A Novel Mechanism of Sulfur Transfer Catalyzed by O-Acetylhomoserine Sulfhydrylase in Methionine Biosynthetic Pathway of Wolinella succinogenes | | Descriptor: | O-ACETYLHOMOSERINE SULFHYDRYLASE | | Authors: | Tran, T.H, Krishnamoorthy, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel mechanism of sulfur transfer catalyzed by O-acetylhomoserine sulfhydrylase in the methionine-biosynthetic pathway of Wolinella succinogenes.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
3J0R
 
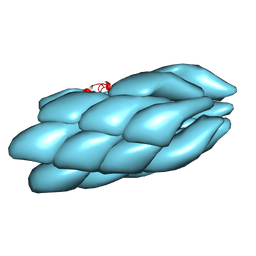 | | Model of a type III secretion system needle based on a 7 Angstrom resolution cryoEM map | | Descriptor: | Protein mxiH | | Authors: | Fujii, T, Cheung, M, Blanco, A, Kato, T, Blocker, A.J, Namba, K. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5H53
 
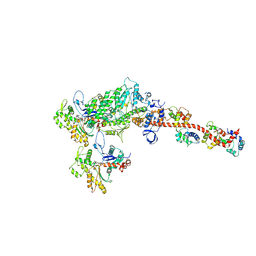 | |
5AX2
 
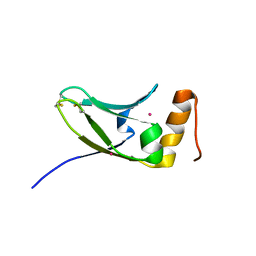 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
6K9Q
 
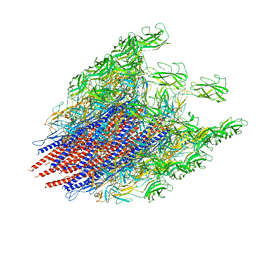 | | Structure of the native supercoiled hook as a universal joint | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Kato, T, Miyata, T, Makino, F, Horvath, P, Namba, K. | | Deposit date: | 2019-06-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the native supercoiled flagellar hook as a universal joint.
Nat Commun, 10, 2019
|
|
7CLR
 
 | | CryoEM structure of S.typhimurium flagellar LP ring | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Yamaguchi, T, Makino, F, Miyata, T, Minamino, T, Kato, T, Namba, K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-02 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the molecular bushing of the bacterial flagellar motor.
Nat Commun, 12, 2021
|
|
