3RO6
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3EUO
 
 | |
3T9W
 
 | | Small laccase from Amycolatopsis sp. ATCC 39116 | | Descriptor: | COPPER (II) ION, HYDROGEN PEROXIDE, NICKEL (II) ION, ... | | Authors: | Lukk, T, Majumdar, S, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Roles of small laccases from Streptomyces in lignin degradation.
Biochemistry, 53, 2014
|
|
3F8T
 
 | |
3OU6
 
 | | DhpI-SAM complex | | Descriptor: | S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, SULFATE ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU7
 
 | | DhpI-SAM-HEP complex | | Descriptor: | (2-hydroxyethyl)phosphonic acid, S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, ... | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3SZY
 
 | |
3T01
 
 | |
3SZZ
 
 | |
3T00
 
 | |
3OU2
 
 | | DhpI-SAH complex structure | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OO3
 
 | |
3DFM
 
 | |
3DFF
 
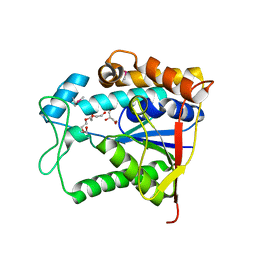 | | The crystal structure of teicoplanin pseudoaglycone deacetylase Orf2 | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Teicoplanin pseudoaglycone deacetylases Orf2, ... | | Authors: | Zou, Y, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of lipoglycopeptide antibiotic deacetylases: implications for the biosynthesis of a40926 and teicoplanin.
Chem.Biol., 15, 2008
|
|
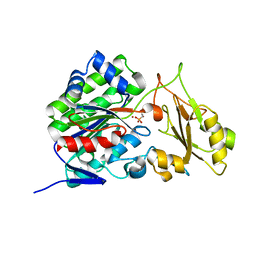
3T02
 
 | |
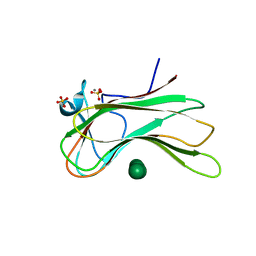
3OEB
 
 | |
3OEA
 
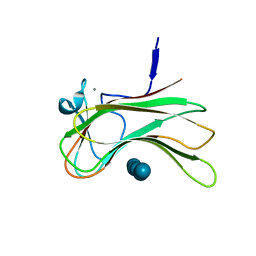 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
3SMA
 
 | |
3TLB
 
 | |
3TLE
 
 | |
3T33
 
 | | Crystal Structure of Arabidopsis GCR2 | | Descriptor: | ACETATE ION, G protein coupled receptor, ZINC ION | | Authors: | Chen, J.-H, Guo, J, Chen, J.-G, Nair, S.K. | | Deposit date: | 2011-07-24 | | Release date: | 2013-04-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Arabidopsis GCR2 Identifies a Novel Clade of Lantibiotic Cyclase-Like Proteins
To be Published
|
|
3TLC
 
 | |
3TLY
 
 | |
3M6I
 
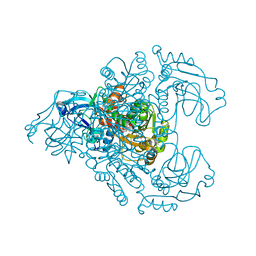 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
3TLA
 
 | |
