5M86
 
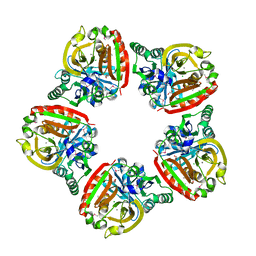 | | Crystal Structure of the Thermoplasma acidophilum Protein Ta1207 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Pathare, G.R, Nagy, I, Bracher, A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-06-14 | | 最終更新日 | 2017-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the Thermoplasma acidophilum protein Ta1207.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7OI3
 
 | | Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex | | 分子名称: | Cetacean morbillivirus nucleoprotein, poly-A 6-mer | | 著者 | Zinzula, L, Beck, F, Klumpe, S, Bohn, S, Pfeifer, G, Bollschweiler, D, Nagy, I, Plitzko, J.M, Baumeister, W. | | 登録日 | 2021-05-11 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of the cetacean morbillivirus nucleoprotein-RNA complex.
J.Struct.Biol., 213, 2021
|
|
6ZCO
 
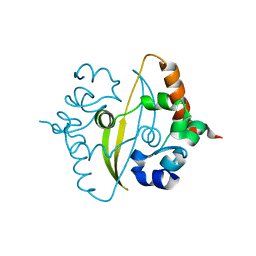 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | 分子名称: | Nucleoprotein | | 著者 | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.361 Å) | | 主引用文献 | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6YUN
 
 | |
2FSJ
 
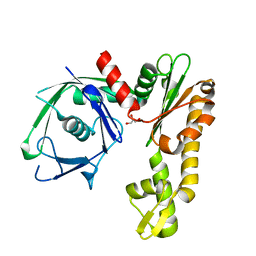 | | Crystal structure of Ta0583, an archaeal actin homolog, native data | | 分子名称: | GLYCEROL, hypothetical protein Ta0583 | | 著者 | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | 登録日 | 2006-01-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2FSK
 
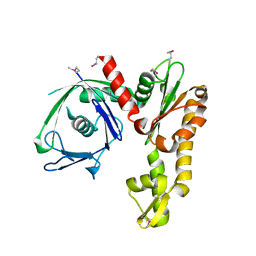 | | Crystal structure of Ta0583, an archaeal actin homolog, SeMet data | | 分子名称: | hypothetical protein Ta0583 | | 著者 | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | 登録日 | 2006-01-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2FSN
 
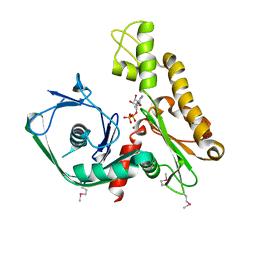 | | Crystal structure of Ta0583, an archaeal actin homolog, complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein Ta0583 | | 著者 | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | 登録日 | 2006-01-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
6FVW
 
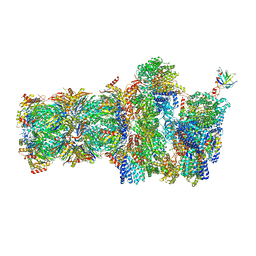 | | 26S proteasome, s4 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVV
 
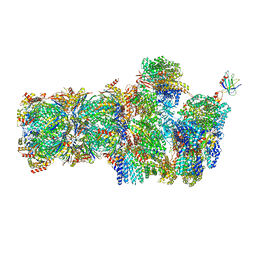 | | 26S proteasome, s3 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
1Q5R
 
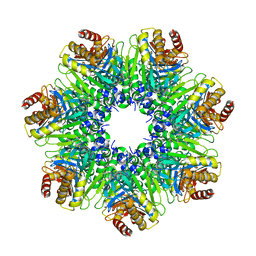 | | The Rhodococcus 20S proteasome with unprocessed pro-peptides | | 分子名称: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | 著者 | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | 登録日 | 2003-08-08 | | 公開日 | 2003-12-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
1Q5Q
 
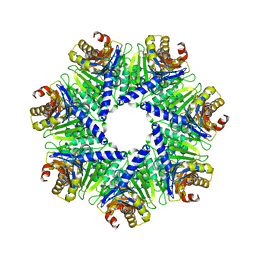 | | The Rhodococcus 20S proteasome | | 分子名称: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | 著者 | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | 登録日 | 2003-08-08 | | 公開日 | 2003-12-16 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
6FVU
 
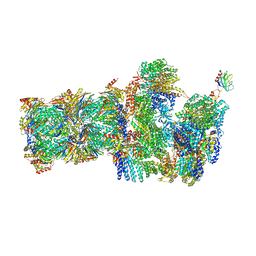 | | 26S proteasome, s2 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVT
 
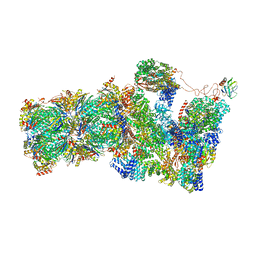 | | 26S proteasome, s1 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVY
 
 | | 26S proteasome, s6 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVX
 
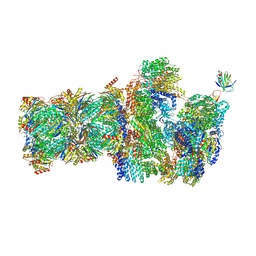 | | 26S proteasome, s5 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6GBO
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Ebola virus | | 分子名称: | Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6GBP
 
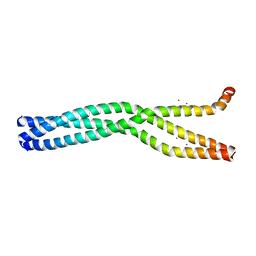 | | Crystal Structure of the oligomerization domain of VP35 from Ebola virus, mercury derivative | | 分子名称: | MERCURY (II) ION, Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6GBQ
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Reston virus | | 分子名称: | Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6GBR
 
 | | Crystal Structure of the oligomerization domain of VP35 from Reston virus, mercury derivative | | 分子名称: | MERCURIBENZOIC ACID, Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
