4S33
 
 | | ERK2 R65S mutant complexed with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
1O7A
 
 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
1O9K
 
 | | Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F peptide | | Descriptor: | RETINOBLASTOMA-ASSOCIATED PROTEIN, TRANSCRIPTION FACTOR E2F1 | | Authors: | Xiao, B, Spencer, J, Clements, A, Ali-Khan, N, Mittnacht, S, Broceno, C, Burghammer, M, Perrakis, A, Marmorstein, R, Gamblin, S.J. | | Deposit date: | 2002-12-16 | | Release date: | 2003-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Retinoblastoma Tumor Suppressor Protein Bound to E2F and the Molecular Basis of its Regulation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4TQ2
 
 | | Structure of S-type Phycobiliprotein Lyase CPES from Guillardia theta | | Descriptor: | HEXANE-1,6-DIOL, Putative phycoerythrin lyase | | Authors: | Gasper, R, Overkamp, K.E, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the Biosynthesis and Assembly of Cryptophycean Phycobiliproteins.
J.Biol.Chem., 289, 2014
|
|
1NMY
 
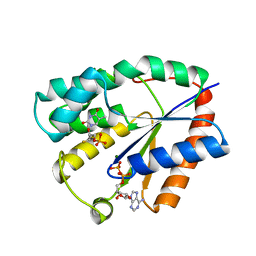 | | Crystal structure of human thymidylate kinase with FLTMP and AppNHp | | Descriptor: | 3'-FLUORO-3'-DEOXYTHYMIDINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Segura-Pena, D, Meier, C, Veit, T, Monnerjahn, M, Konrad, M, Lavie, A. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human thymidylate kinase in complex with prodrugs:
implications for the structure-based design of novel compounds
Biochemistry, 42, 2003
|
|
2RSC
 
 | |
2RSI
 
 | |
4TXV
 
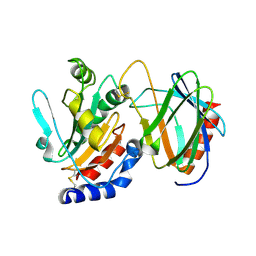 | | Crystal structure of the mixed disulfide intermediate between thioredoxin-like TlpAs(C110S) and subunit II of cytochrome c oxidase CoxBPD (C233S) | | Descriptor: | Cytochrome c oxidase subunit 2, Thiol:disulfide interchange protein TlpA | | Authors: | Quade, N, Abicht, H.K, Hennecke, H, Glockshuber, R. | | Deposit date: | 2014-07-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
1P6V
 
 | | Crystal structure of the tRNA domain of transfer-messenger RNA in complex with SmpB | | Descriptor: | 45-MER, SsrA-binding protein | | Authors: | Gutmann, S, Haebel, P.W, Metzinger, L, Sutter, M, Felden, B, Ban, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB
Nature, 424, 2003
|
|
1P9O
 
 | | Crystal Structure of Phosphopantothenoylcysteine Synthetase | | Descriptor: | Phosphopantothenoylcysteine synthetase, SULFATE ION | | Authors: | Manoj, N, Strauss, E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-05-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human phosphopantothenoylcysteine synthetase at 2.3 A resolution.
Structure, 11, 2003
|
|
1OQ0
 
 | |
4U1D
 
 | |
1OXX
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
1OIB
 
 | |
5NOA
 
 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | Family 88 glycosyl hydrolase | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
1OGP
 
 | | The crystal structure of plant sulfite oxidase provides insight into sulfite oxidation in plants and animals | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(VI), CESIUM ION, GLYCEROL, ... | | Authors: | Schrader, N, Fischer, K, Theis, K, Mendel, R.R, Schwarz, G, Kisker, C. | | Deposit date: | 2003-05-08 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Plant Sulfite Oxidase Provides Insights Into Sulfite Oxidation in Plants and Animals
Structure, 11, 2003
|
|
5NNY
 
 | |
4U4Y
 
 | | Crystal structure of Pactamycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U1C
 
 | |
1OZJ
 
 | | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | | Descriptor: | SMAD 3, Smad binding element, ZINC ION | | Authors: | Chai, J, Wu, J.-W, Yan, N, Massague, J, Pavletich, N.P, Shi, Y. | | Deposit date: | 2003-04-09 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding.
J.Biol.Chem., 278, 2003
|
|
5MB3
 
 | |
4U98
 
 | | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway (AppCp complex) | | Descriptor: | MAGNESIUM ION, Maltokinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Fraga, J, Empadinhas, N, Pereira, P.J.B, Macedo-Ribeiro, S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway.
Sci Rep, 5, 2015
|
|
1P1Q
 
 | |
1OM6
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5mM EDTA (2 MONTHS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1P1U
 
 | |
