2E22
 
 | | Crystal structure of xanthan lyase in complex with mannose | | Descriptor: | Xanthan lyase, alpha-D-mannopyranose | | Authors: | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
7DOU
 
 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7DN2
 
 | | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Cement protein gp15, Major structural protein ORF14 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
3VLU
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with saturated trimannuronate | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VLV
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding ptotein AlgQ1 in complex with unsaturated triguluronate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlgQ1, CALCIUM ION | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3A1K
 
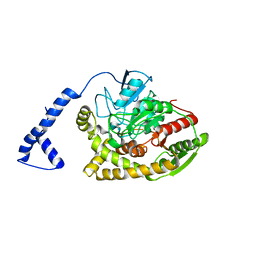 | | Crystal structure of Rhodococcus sp. N771 Amidase | | Descriptor: | Amidase | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
7F2P
 
 | | The head structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16, KHP40 MCP | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2021-06-13 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
3VWO
 
 | |
3WIW
 
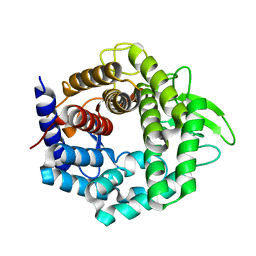 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3A1I
 
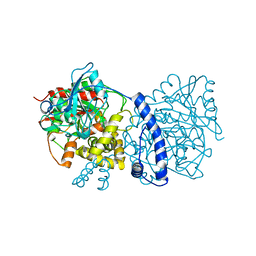 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | Descriptor: | Amidase, BENZAMIDE | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
2Z42
 
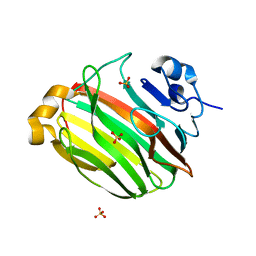 | | Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Recognition by Family 7 Alginagte Lyase from Sphingomonas sp. A1
To be published
|
|
2ZAB
 
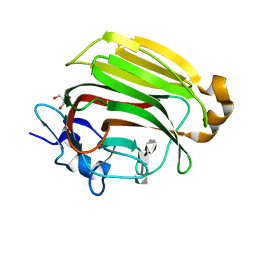 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | Descriptor: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | Descriptor: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2Z8R
 
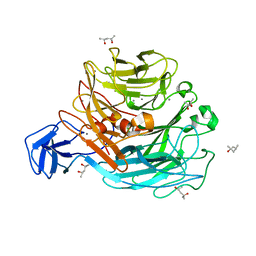 | | Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2ZAA
 
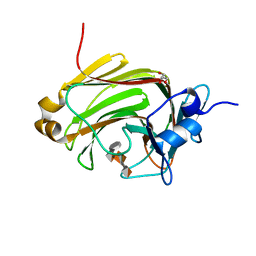 | | Crystal Structure of Family 7 Alginate Lyase A1-II' H191N/Y284F in Complex with Substrate (GGMG) | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase, GLYCEROL | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
7E55
 
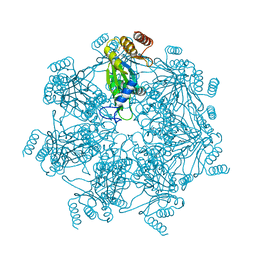 | | Cryo-EM structure of alpha 7 homo-tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Song, C, Murata, K. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural Fluctuations of the Human Proteasome alpha 7 Homo-Tetradecamer Double Ring Imply the Proteasomal alpha-Ring Assembly Mechanism.
Int J Mol Sci, 22, 2021
|
|
2ZYC
 
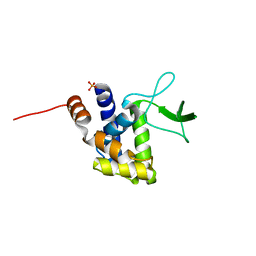 | | Crystal structure of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | PHOSPHATE ION, Peptidoglycan hydrolase FlgJ | | Authors: | Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3A0N
 
 | | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
2ZA9
 
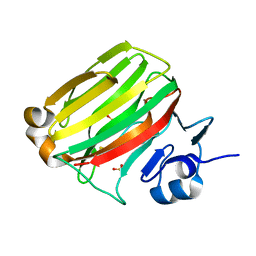 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
3AFO
 
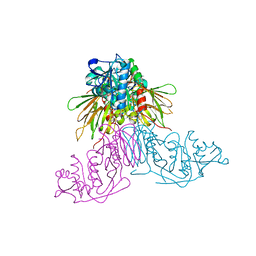 | | Crystal Structure of Yeast NADH Kinase complexed with NADH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NADH kinase POS5 | | Authors: | Ando, T, Ohashi, K, Ochiai, A, Miyagi, H, Kawai, S, Mikami, B, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural factor conferring NADH kinase activity on yeast mitochondrial Pos5
To be Published
|
|
2ZUY
 
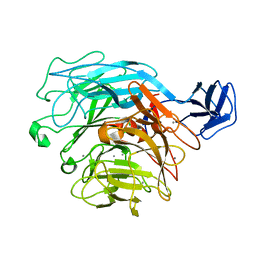 | | Crystal structure of exotype rhamnogalacturonan lyase YesX | | Descriptor: | CALCIUM ION, YesX protein | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
2Z8S
 
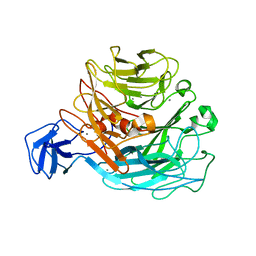 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2ZUX
 
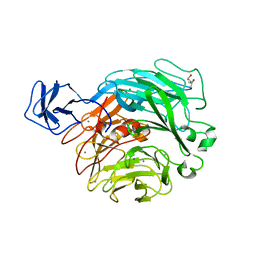 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein, ... | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
3AFN
 
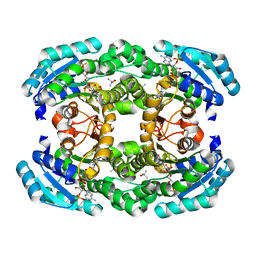 | | Crystal structure of aldose reductase A1-R complexed with NADP | | Descriptor: | Carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TERTIARY-BUTYL ALCOHOL | | Authors: | Takase, R, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1
Biochim.Biophys.Acta, 1804, 2010
|
|
2ZBL
 
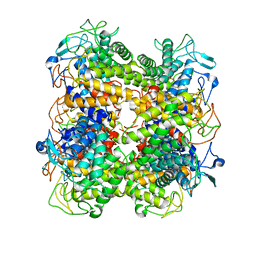 | | Functional annotation of Salmonella enterica yihS-encoded protein | | Descriptor: | Putative isomerase, beta-D-mannopyranose | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
