3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MMH
 
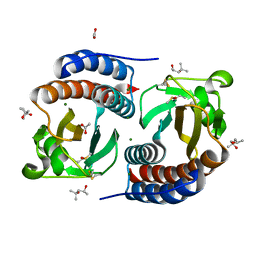 | | X-ray structure of free methionine-R-sulfoxide reductase from neisseria meningitidis in complex with its substrate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Gruez, A, Libiad, M, Boschi-Muller, S, Branlant, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Free Methionine-R-sulfoxide Reductase from Neisseria meningitidis.
J.Biol.Chem., 285, 2010
|
|
2K0R
 
 | | Solution structure of the C103S mutant of the N-terminal Domain of DsbD from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Quinternet, M, Selme, L, Tsan, P, Beaufils, C, Jacob, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the cysteine 103 to serine mutant of the N-terminal domain of DsbD from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2K9F
 
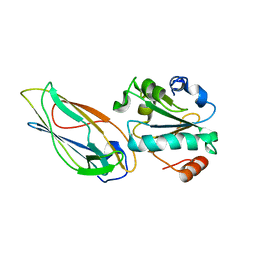 | | Structural features of the complex between the DsbD N-terminal and the PilB N-terminal domains from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein dsbD, Thioredoxin | | Authors: | Quinternet, M, Tsan, P, Selme, L, Jacob, C, Boschi-Muller, S, Branlant, G, Cung, M. | | Deposit date: | 2008-10-09 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Formation of the complex between DsbD and PilB N-terminal domains from Neisseria meningitidis necessitates an adaptability of nDsbD.
Structure, 17, 2009
|
|
2JZS
 
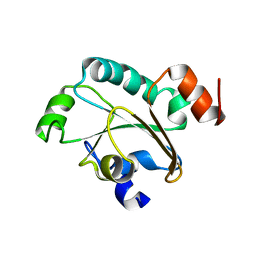 | | Solution structure of the reduced form of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2JZR
 
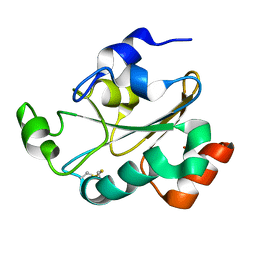 | | Solution structure of the oxidized form (Cys67-Cys70) of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
1NQA
 
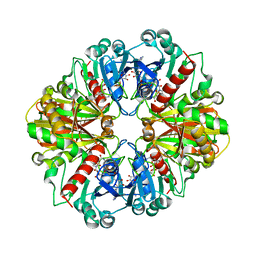 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQO
 
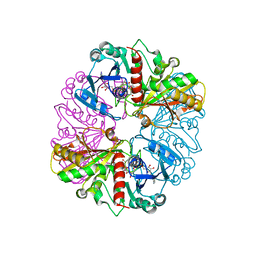 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQ5
 
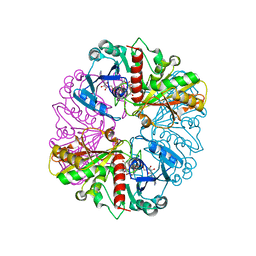 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
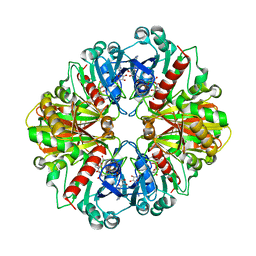 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
2J89
 
 | | Functional and structural aspects of poplar cytosolic and plastidial type A methionine sulfoxide reductases | | Descriptor: | BETA-MERCAPTOETHANOL, METHIONINE SULFOXIDE REDUCTASE A | | Authors: | Rouhier, N, Kauffmann, B, Tete-Favier, F, Palladino, P, Gans, P, Branlant, G, Jacquot, J.P, Boschi-Muller, S. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Aspects of Poplar Cytosolic and Plastidial Type a Methionine Sulfoxide Reductases
J.Biol.Chem., 282, 2007
|
|
1FF3
 
 | | STRUCTURE OF THE PEPTIDE METHIONINE SULFOXIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE, SULFATE ION | | Authors: | Tete-Favier, F, Cobessi, D, Boschi-Muller, S, Azza, S, Branlant, G, Aubry, A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 A resolution.
Structure Fold.Des., 8, 2000
|
|
2FY6
 
 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
5T4U
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with a quinolinone ligand | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, NITRATE ION, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
5T4V
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-48 ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-N-(7-methoxy-1,4-dimethyl-2-oxo-1,2-dihydroquinolin-6-yl)benzene-1-sulfonamide, FORMIC ACID, ... | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
2IEM
 
 | | Solution structure of an oxidized form (Cys51-Cys198) of E. coli Methionine Sulfoxide Reductase A (MsrA) | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Coudevylle, N, Antoine, M, Bouguet-Bonnet, S, Mutzenhardt, P, Boschi-Muller, S, Branlant, G, Cung, M.T. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Reduced Form and an Oxidized Form of E. coli Methionine Sulfoxide Reductase A (MsrA): Structural Insight of the MsrA Catalytic Cycle.
J.Mol.Biol., 366, 2007
|
|
2XF8
 
 | | Structure of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli in complex with a NAD cofactor analog (3-Chloroacetyl adenine pyridine dinucleotide) and sulfate anion | | Descriptor: | 3-(CHLOROACETYL) PYRIDINE ADENINE DINUCLEOTIDE, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5J
 
 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
1CW8
 
 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO (mA-R)REGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 6 STRUCTURES | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, T. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CT6
 
 | | SOLUTION STRUCTURE OF CGGIRGERG IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 11 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-19 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CS9
 
 | | SOLUTION STRUCTURE OF CGGIRGERA IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1BFW
 
 | | RETRO-INVERSO ANALOGUE OF THE G-H LOOP OF VP1 IN FOOT-AND-MOUTH-DISEASE (FMD) VIRUS, NMR, 10 STRUCTURES | | Descriptor: | VP1 PROTEIN | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-22 | | Release date: | 1999-01-13 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
2X5K
 
 | | Structure of an active site mutant of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, ... | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
1BCV
 
 | | SYNTHETIC PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS, NMR, 10 STRUCTURES | | Descriptor: | PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-03 | | Release date: | 1998-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1CVQ
 
 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO MGREGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-24 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
