5EVY
 
 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5B5J
 
 | | Hen egg white lysozyme with boron tracedrug UTX-97 | | Descriptor: | 2-cyano-3-((6-(((2-((2-cyanoethyl)(borocaptate-10B)sulfonio)acetyl)carbamoyl)oxy)hexyl)amino)quinoxaline 1,4-dioxide, Lysozyme C, SODIUM ION | | Authors: | Morimoto, Y. | | Deposit date: | 2016-05-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural Insight Into Protein Binding of Boron Tracedrug UTX-97 Revealed by the Co-Crystal Structure With Lysozyme at 1.26 angstrom Resolution.
J Pharm Sci, 105, 2016
|
|
2CYM
 
 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1993-09-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
2DU2
 
 | | Crystal Structure Analysis of the L-Lactate Oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Morimoto, Y. | | Deposit date: | 2006-07-19 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of L-lactate oxidase from Aerococcus viridans at 2.1A resolution reveals the mechanism of strict substrate recognition
Biochem.Biophys.Res.Commun., 350, 2006
|
|
2E77
 
 | | Crystal structure of L-lactate oxidase with pyruvate complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Morimoto, Y. | | Deposit date: | 2007-01-06 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study on the interaction of L-lactate oxidase with pyruvate at 1.9 Angstrom resolution.
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2DXM
 
 | | Neutron Structure Analysis of Deoxy Human Hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y. | | Deposit date: | 2006-08-28 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Protonation states of buried histidine residues in human deoxyhemoglobin revealed by neutron crystallography.
J.Am.Chem.Soc., 129, 2007
|
|
3AB5
 
 | |
7F21
 
 | | L-lactate oxidase with D-lactate | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, LACTIC ACID | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F22
 
 | | L-lactate oxidase with pyruvate | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, PYRUVIC ACID | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F20
 
 | | L-lactate oxidase with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, FLAVIN MONONUCLEOTIDE, L-lactate oxidase | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F1Y
 
 | | L-lactate oxidase without substrate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
1QVC
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
3KMF
 
 | |
1IRU
 
 | | Crystal Structure of the mammalian 20S proteasome at 2.75 A resolution | | Descriptor: | 20S proteasome, MAGNESIUM ION | | Authors: | Unno, M, Mizushima, T, Morimoto, Y, Tomisugi, Y, Tanaka, K, Yasuoka, N, Tsukihara, T. | | Deposit date: | 2001-10-24 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of the mammalian 20S proteasome at 2.75 A resolution.
Structure, 10, 2002
|
|
6K8G
 
 | | H/D exchanged Hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen/deuterium exchange behavior in tetragonal hen egg-white lysozyme crystals affected by solution state.
J.Appl.Crystallogr., 53, 2020
|
|
1EEX
 
 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Shibata, N, Masuda, J, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-04 | | Release date: | 2001-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1EGV
 
 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSELLA OXYTOCA UNDER THE ILLUMINATED CONDITION. | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-17 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1IWB
 
 | | Crystal structure of diol dehydratase | | Descriptor: | COBALAMIN, DIOL DEHYDRATASE alpha chain, DIOL DEHYDRATASE beta chain, ... | | Authors: | Shibata, N, Masuda, J, Morimoto, Y, Yasuoka, N, Toraya, T. | | Deposit date: | 2002-05-01 | | Release date: | 2003-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-induced conformational change of a coenzyme B12-dependent enzyme: crystal structure of the substrate-free form of diol dehydratase
Biochemistry, 41, 2002
|
|
1EGM
 
 | | CRYSTAL STRUCTURE OF DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX AT 100K. | | Descriptor: | CYANOCOBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-15 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1X0P
 
 | | Structure of a cyanobacterial BLUF protein, Tll0078 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, hypothetical protein Tll0078 | | Authors: | Kita, A, Okajima, K, Morimoto, Y, Ikeuchi, M, Miki, K. | | Deposit date: | 2005-03-27 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Cyanobacterial BLUF Protein, Tll0078, Containing a Novel FAD-binding Blue Light Sensor Domain
J.Mol.Biol., 349, 2005
|
|
1WQ5
 
 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
5B05
 
 | | Lysozyme (control experiment) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5B07
 
 | | Lysozyme (denatured by DCl and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5B06
 
 | | Lysozyme (denatured by NaOD and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
2CVC
 
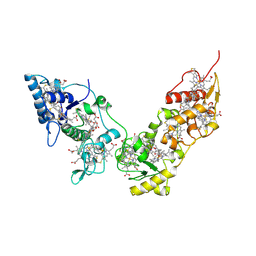 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
