1EQ4
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQ5
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-04 | | 公開日 | 2000-04-19 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1ENI
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | 分子名称: | ENDONUCLEASE V | | 著者 | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | 登録日 | 1994-08-08 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENK
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | 分子名称: | ENDONUCLEASE V | | 著者 | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | 登録日 | 1994-08-08 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1EWK
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | 著者 | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | 登録日 | 2000-04-26 | | 公開日 | 2000-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | 分子名称: | DrP35 | | 著者 | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | 登録日 | 2006-03-07 | | 公開日 | 2006-12-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
1XPA
 
 | | SOLUTION STRUCTURE OF THE DNA-AND RPA-BINDING DOMAIN OF THE HUMAN REPAIR FACTOR XPA, NMR, 1 STRUCTURE | | 分子名称: | XPA, ZINC ION | | 著者 | Ikegami, T, Kuraoka, I, Saijo, M, Kodo, N, Kyogoku, Y, Morikawa, K, Tanaka, K, Shirakawa, M. | | 登録日 | 1998-07-06 | | 公開日 | 1999-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA.
Nat.Struct.Biol., 5, 1998
|
|
1AKL
 
 | | ALKALINE PROTEASE FROM PSEUDOMONAS AERUGINOSA IFO3080 | | 分子名称: | ALKALINE PROTEASE, CALCIUM ION, ZINC ION | | 著者 | Miyatake, H, Hata, Y, Fujii, T, Hamada, K, Morihara, K, Katsube, Y. | | 登録日 | 1995-09-16 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the unliganded alkaline protease from Pseudomonas aeruginosa IFO3080 and its conformational changes on ligand binding.
J.Biochem.(Tokyo), 118, 1995
|
|
1LAW
 
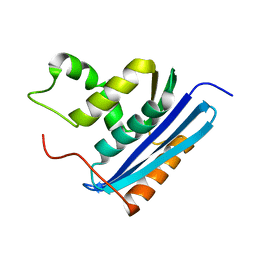 | |
1LAV
 
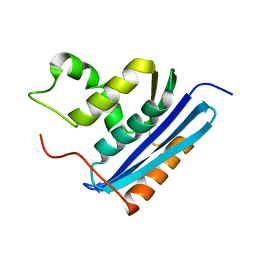 | |
2QUL
 
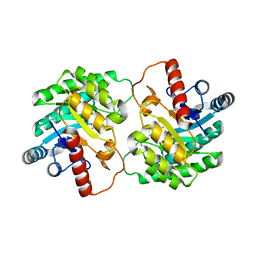 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii at 1.79 A resolution | | 分子名称: | D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
3ITY
 
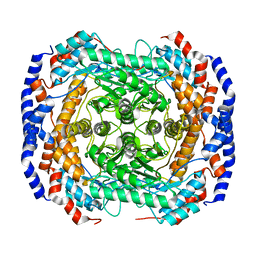 | | Metal-free form of Pseudomonas stutzeri L-rhamnose isomerase | | 分子名称: | L-rhamnose isomerase | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
2QUM
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii with D-tagatose | | 分子名称: | D-tagatose, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
1DQ3
 
 | |
2OU4
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii | | 分子名称: | D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-02-09 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUN
 
 | | Crystal Structure of D-tagatose 3-epimerase from Pseudomonas cichorii in Complex with D-fructose | | 分子名称: | D-fructose, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
1EWV
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | 分子名称: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | 著者 | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | 登録日 | 2000-04-27 | | 公開日 | 2000-12-18 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | 分子名称: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
1AK7
 
 | | DESTRIN, NMR, 20 STRUCTURES | | 分子名称: | DESTRIN | | 著者 | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | 登録日 | 1997-05-29 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1IQP
 
 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | 著者 | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | 登録日 | 2001-07-24 | | 公開日 | 2001-09-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
2DSO
 
 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | 分子名称: | CALCIUM ION, Drp35, GLYCEROL | | 著者 | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | 登録日 | 2006-07-04 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
4XSL
 
 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | 分子名称: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
1IXS
 
 | | Structure of RuvB complexed with RuvA domain III | | 分子名称: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | 著者 | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1IXR
 
 | | RuvA-RuvB complex | | 分子名称: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | 著者 | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
