3MOO
 
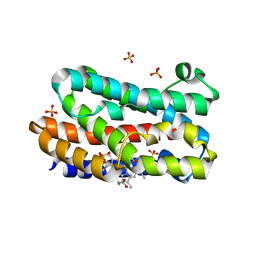 | | Crystal structure of the HmuO, heme oxygenase from Corynebacterium diphtheriae, in complex with azide-bound verdoheme | | 分子名称: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, AZIDE ION, Heme oxygenase, ... | | 著者 | Omori, K, Matsui, T, Unno, M, Ikeda-Saito, M. | | 登録日 | 2010-04-22 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Enzymatic ring-opening mechanism of verdoheme by the heme oxygenase: a combined X-ray crystallography and QM/MM study.
J.Am.Chem.Soc., 132, 2010
|
|
6AK3
 
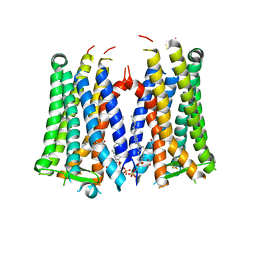 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | 著者 | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | 登録日 | 2018-08-29 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
1X0G
 
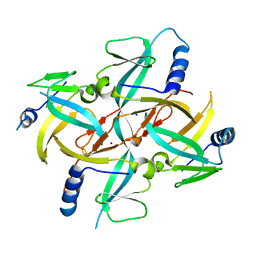 | | Crystal Structure of IscA with the [2Fe-2S] cluster | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, IscA, SODIUM ION | | 著者 | Morimoto, K, Yamashita, E, Kondou, Y, Lee, S.J, Tsukihara, T, Nakai, M. | | 登録日 | 2005-03-22 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Asymmetric IscA Homodimer with an Exposed [2Fe-2S] Cluster Suggests the Structural Basis of the Fe-S Cluster Biosynthetic Scaffold.
J.Mol.Biol., 360, 2006
|
|
2D0P
 
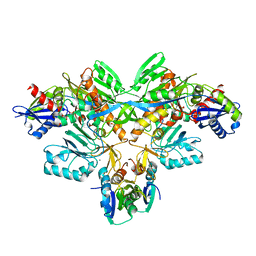 | | Structure of diol dehydratase-reactivating factor in nucleotide free form | | 分子名称: | CALCIUM ION, SULFATE ION, diol dehydratase-reactivating factor large subunit, ... | | 著者 | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | 登録日 | 2005-08-05 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
2D0O
 
 | | Structure of diol dehydratase-reactivating factor complexed with ADP and Mg2+ | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | 登録日 | 2005-08-05 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
1HJR
 
 | |
4XSL
 
 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | 分子名称: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
1HJP
 
 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | 分子名称: | RUVA | | 著者 | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | 登録日 | 1997-08-21 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
3M0X
 
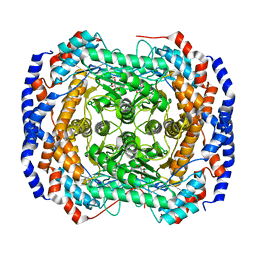 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | 分子名称: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0H
 
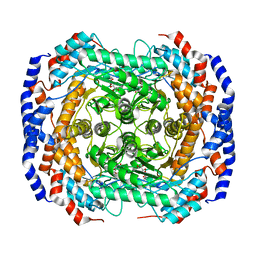 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with L-rhamnose | | 分子名称: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0Y
 
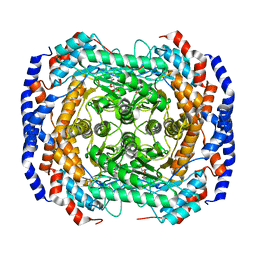 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329A in complex with L-rhamnose | | 分子名称: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0L
 
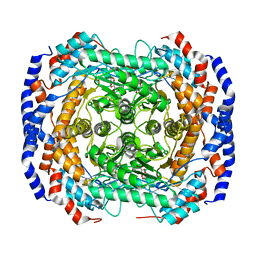 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-psicose | | 分子名称: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0V
 
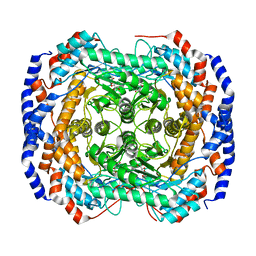 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with L-rhamnose | | 分子名称: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0M
 
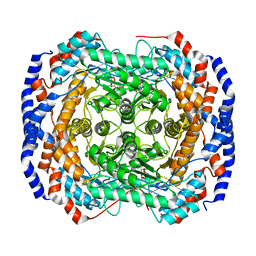 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-allose | | 分子名称: | D-ALLOSE, L-rhamnose isomerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | 登録日 | 2010-03-03 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
8SLJ
 
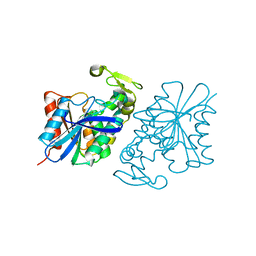 | |
1WP9
 
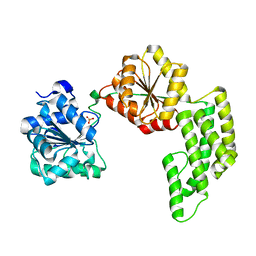 | | Crystal structure of Pyrococcus furiosus Hef helicase domain | | 分子名称: | ATP-dependent RNA helicase, putative, PHOSPHATE ION | | 著者 | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | 登録日 | 2004-08-31 | | 公開日 | 2005-02-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing
Structure, 13, 2005
|
|
1A99
 
 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | 分子名称: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | 著者 | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | 登録日 | 1998-04-17 | | 公開日 | 1998-10-21 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | 分子名称: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
1T88
 
 | | Crystal Structure of the Ferrous Cytochrome P450cam (C334A) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | 著者 | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | 登録日 | 2004-05-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T87
 
 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam (C334A) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CARBON MONOXIDE, ... | | 著者 | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | 登録日 | 2004-05-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
2AEN
 
 | | Crystal structure of the rotavirus strain DS-1 VP8* core | | 分子名称: | ETHANOL, GLYCEROL, Outer capsid protein VP4, ... | | 著者 | Monnier, N, Higo-Moriguchi, K, Sun, Z.-Y.J, Prasad, B.V.V, Taniguchi, K, Dormitzer, P.R. | | 登録日 | 2005-07-22 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | High-resolution molecular and antigen structure of the VP8*
core of a sialic acid-independent human rotavirus strain
J.Virol., 80, 2006
|
|
1T85
 
 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam Mutant (L358P/C334A) | | 分子名称: | CAMPHOR, CARBON MONOXIDE, Cytochrome P450-cam, ... | | 著者 | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | 登録日 | 2004-05-11 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1C46
 
 | |
1C43
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | 分子名称: | PROTEIN (HUMAN LYSOZYME), SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 1999-08-03 | | 公開日 | 1999-08-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C45
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | 分子名称: | PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 1999-08-03 | | 公開日 | 1999-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
