2EWI
 
 | |
2EWU
 
 | |
7XVJ
 
 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
7Y3V
 
 | | Crystal structure of CdpNPT in complex with harmane | | Descriptor: | 1-methyl-9H-pyrido[3,4-b]indole, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-06-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic potential of a fungal indole prenyltransferase toward beta-carbolines, harmine and harman, and their prenylation effects on antibacterial activity.
J.Biosci.Bioeng., 134, 2022
|
|
3NBS
 
 | | Crystal structure of dimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBT
 
 | | Crystal structure of trimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8XDU
 
 | |
8XE4
 
 | | norbelladine 4'-O-methyltransferase complexed with Mg, SAH, and norbelladine | | Descriptor: | GLYCEROL, MAGNESIUM ION, Norbelladine, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDP
 
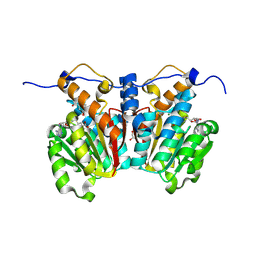 | | O-methyltransferase from Lycoris longituba complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protocatechuic aldehyde, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDY
 
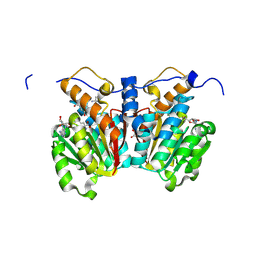 | | O-methyltransferase from Lycoris longituba M52A variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDQ
 
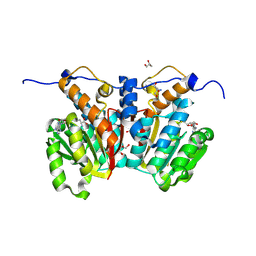 | | O-methyltransferase from Lycoris aurea complexed with Mg and SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDT
 
 | |
8XDO
 
 | |
8XDR
 
 | | O-methyltransferase from Lycoris aurea complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDS
 
 | |
8XE5
 
 | | norbelladine 4'-O-methyltransferase complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Norbelladine 4'-O-methyltransferase, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XE3
 
 | |
8XDV
 
 | |
8XE0
 
 | | O-methyltransferase from Lycoris longituba M52W variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | MAGNESIUM ION, Protocatechuic aldehyde, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XE2
 
 | | O-methyltransferase from Lycoris longituba D230A variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protocatechuic aldehyde, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
2G7M
 
 | | Crystal structure of B. fragilis N-succinylornithine transcarbamylase P90E mutant complexed with carbamoyl phosphate and N-acetylnorvaline | | Descriptor: | N-ACETYL-L-NORVALINE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, SULFATE ION, ... | | Authors: | Shi, D, Yu, X, Roth, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2006-02-28 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
2FMY
 
 | |
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
7V5Q
 
 | | The dimeric structure of G80A/H81A/L137E myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
7V5R
 
 | | The dimeric structure of G80A/H81A/L137D myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
