6TG2
 
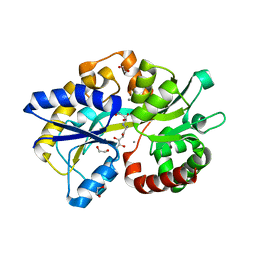 | | Structure of the PBP/SBP MotA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFS
 
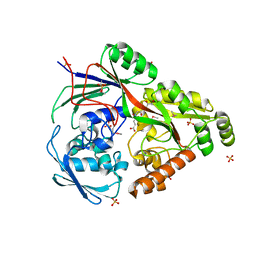 | | Structure in P3212 form of the PBP/SBP MoaA in complex with glucopinic acid from A.tumefacien R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, ABC transporter substrate-binding protein, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
4MLA
 
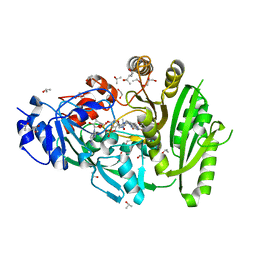 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin oxidase 2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
4ML8
 
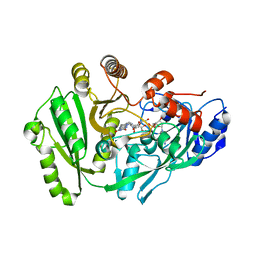 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | Cytokinin oxidase 2, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3IP5
 
 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP7
 
 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP6
 
 | | Structure of Atu2422-GABA receptor in complex with proline | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), PROLINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPC
 
 | | Structure of ATU2422-GABA F77A mutant receptor in complex with leucine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), LEUCINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPA
 
 | | Structure of ATU2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP9
 
 | | Structure of Atu2422-GABA receptor in complex with GABA | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
5L9P
 
 | | Crystal structure of the PBP MotA from A. tumefaciens B6 | | Descriptor: | SULFATE ION, periplasmic binding protein | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2016-06-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
6TG3
 
 | | Crystal Structure of the PBP/SBP MotA in complex with glucopinic acid from A. tumefaciens B6/R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, MotA | | Authors: | Morera, S, Vigouroux, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
8CAY
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Siragu, S. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
1C3J
 
 | | T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, A, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism.
J.Mol.Biol., 292, 1999
|
|
8C6U
 
 | | PBP AccA-G145YG440Q mutant from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH3
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6R
 
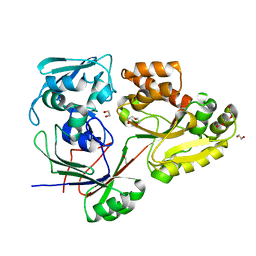 | | PBP AccA from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH1
 
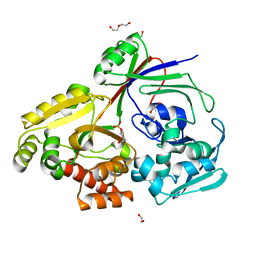 | | PBP AccA from A. vitis S4 in complex with Agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKD
 
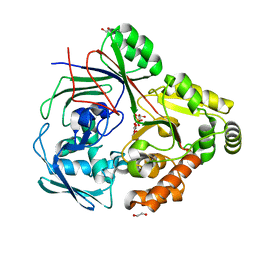 | | PBP AccA from A. fabrum C58 in complex with agrocinopine D-like | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C75
 
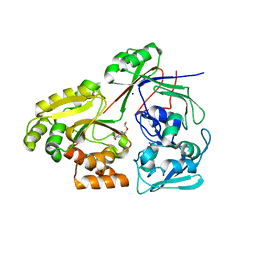 | | PBP AccA from A. tumefaciens Bo542 in apoform 3 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CJU
 
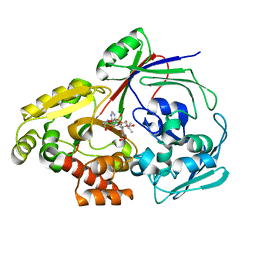 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKO
 
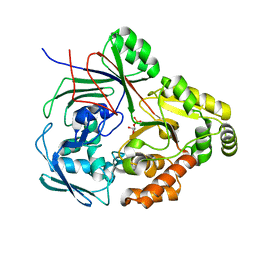 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6W
 
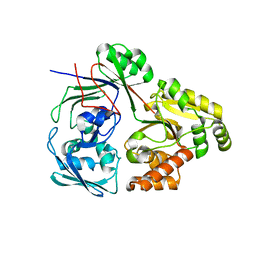 | | PBP AccA from A. tumefaciens Bo542 in apoform 1 | | Descriptor: | Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CAW
 
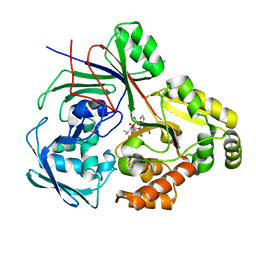 | | PBP AccA from A. tumefaciens Bo542 in complex with agrocin84 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, [(2R,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
