6Z74
 
 | |
6ZA7
 
 | | Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
4EA4
 
 | | Structure of the glycosylase domain of MBD4 bound to 5hmU-containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLZ
 
 | | Structure in C2 form of the PBP AgtB from A.tumefacien R10 in complex with agropinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Agropine permease, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLX
 
 | | Structure of the PBP AgaA in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
1QKJ
 
 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
6HQH
 
 | |
5L9P
 
 | | Crystal structure of the PBP MotA from A. tumefaciens B6 | | Descriptor: | SULFATE ION, periplasmic binding protein | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2016-06-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
1NUE
 
 | | X-RAY STRUCTURE OF NM23 HUMAN NUCLEOSIDE DIPHOSPHATE KINASE B COMPLEXED WITH GDP AT 2 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Morera, S, Lacombe, M.-L, Yingwu, X, Lebras, G, Janin, J. | | Deposit date: | 1995-10-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution.
Structure, 3, 1995
|
|
6I7W
 
 | | Structure of the periplasmic binding protein (PBP) AccA in complex with 2-glucose-2-O-lactic acid phosphate (G2LP) from Agrobacterium fabrum C58 | | Descriptor: | 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-beta-D-glucopyranose, ABC transporter, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of a non-natural glucose-2-phosphate ester able to dupe the acc system of Agrobacterium fabrum.
Org. Biomol. Chem., 17, 2019
|
|
6HM2
 
 | | Structure in P1 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, SODIUM ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6QAK
 
 | |
6QAP
 
 | |
6R5S
 
 | |
6R6K
 
 | | Structure of a FpvC mutant from pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa.
Febs J., 287, 2020
|
|
6R3Z
 
 | |
6R44
 
 | |
6RU4
 
 | |
6QAO
 
 | |
3RX5
 
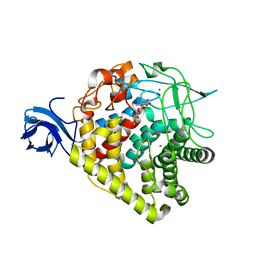 | | structure of AaCel9A in complex with cellotriose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl 4-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, Cellulase, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fortuitous binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidase
Org.Biomol.Chem., 9, 2011
|
|
3RX7
 
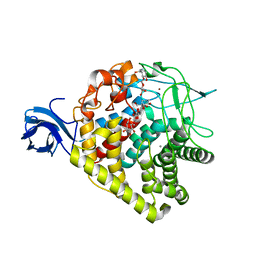 | | Structure of AaCel9A in complex with cellotetraose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
3RX8
 
 | | structure of AaCel9A in complex with cellobiose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
4K1G
 
 | |
4I8Q
 
 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
