1OED
 
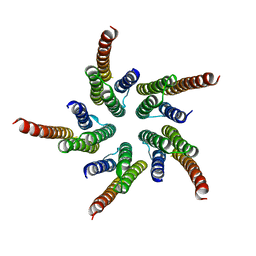 | | STRUCTURE OF ACETYLCHOLINE RECEPTOR PORE FROM ELECTRON IMAGES | | Descriptor: | Acetylcholine receptor beta subunit, Acetylcholine receptor delta subunit, Acetylcholine receptor gamma subunit, ... | | Authors: | Miyazawa, A, Fujiyoshi, Y, Unwin, N. | | Deposit date: | 2003-03-24 | | Release date: | 2003-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and Gating Mechanism of the Acetylcholine Receptor Pore.
Nature, 423, 2003
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
2AT9
 
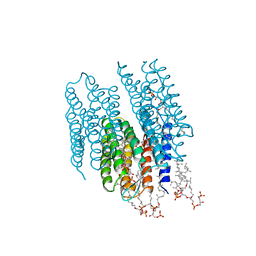 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
1AT9
 
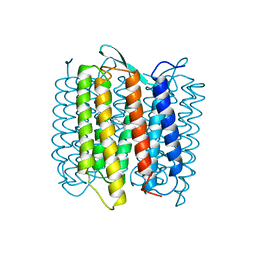 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
