2AT9
 
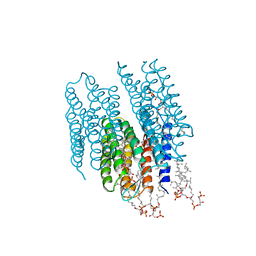 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
6LY9
 
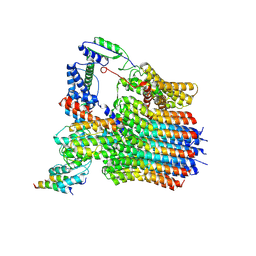 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6LY8
 
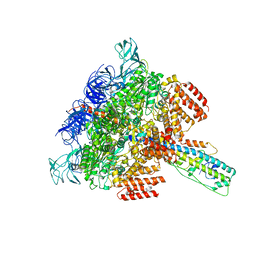 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
1AT9
 
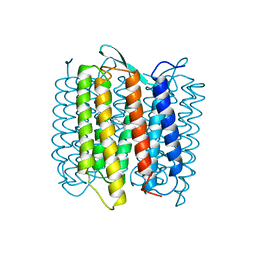 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
1I2H
 
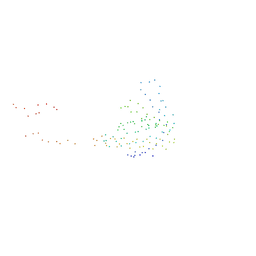 | | CRYSTAL STRUCTURE ANALYSIS OF PSD-ZIP45(HOMER1C/VESL-1L)CONSERVED HOMER 1 DOMAIN | | Descriptor: | PSD-ZIP45(HOMER-1C/VESL-1L) | | Authors: | Irie, K, Nakatsu, T, Mitsuoka, K, Fujiyoshi, Y, Kato, H. | | Deposit date: | 2001-02-09 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Homer 1 Family Conserved Region Reveals the Interaction Between the EVH1 Domain and
Own Proline-rich Motif
J.Mol.Biol., 318, 2002
|
|
7WYI
 
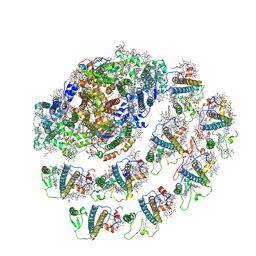 | | Native Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
7WZN
 
 | | PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
2D57
 
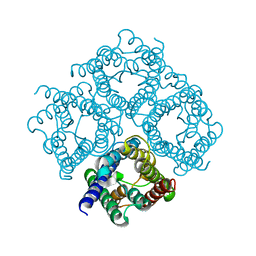 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
1FQY
 
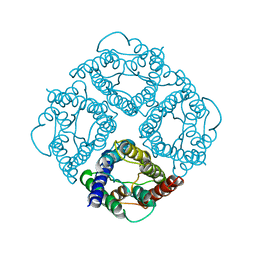 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLO
 
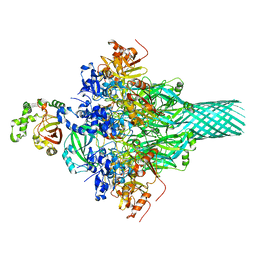 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLW
 
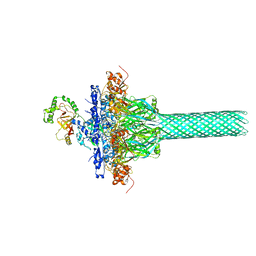 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5Y5Y
 
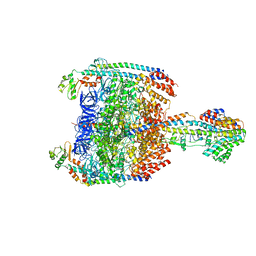 | | V/A-type ATPase/synthase from Thermus thermophilus, peripheral domain, rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-24 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y5X
 
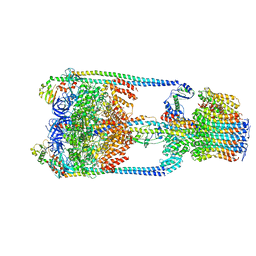 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y5Z
 
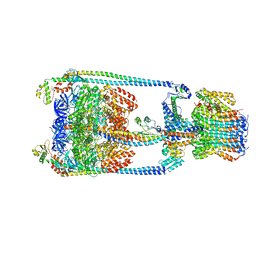 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5Y60
 
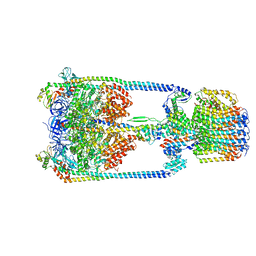 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
8HH2
 
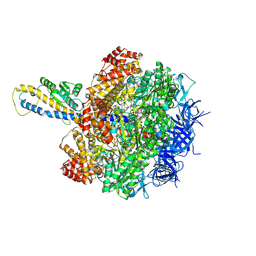 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH4
 
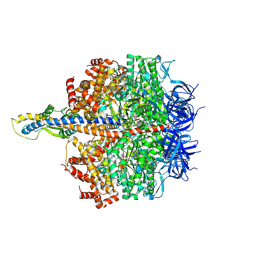 | | F1 domain of FoF1-ATPase from Bacillus PS3,101 degrees, highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH1
 
 | | FoF1-ATPase from Bacillus PS3, 81 degrees, highATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH3
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,90 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8GXX
 
 | | 3 nucleotide-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXU
 
 | | 1 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SULFATE ION, V-type ATP synthase alpha chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXZ
 
 | | 1 sulfate and 1 ATP bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXY
 
 | | 2 sulfate-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | SULFATE ION, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXW
 
 | | 2 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
