6J92
 
 | |
6ZMV
 
 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAO
 
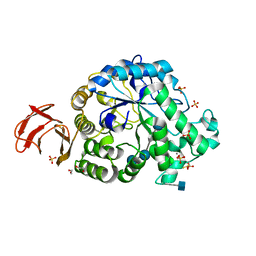 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
8B2E
 
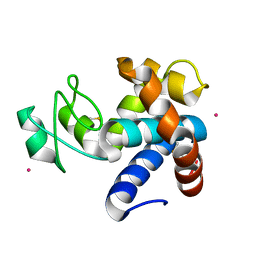 | | Muramidase from Kionochaeta sp natural catalytic core | | Descriptor: | CADMIUM ION, Muramidase | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2G
 
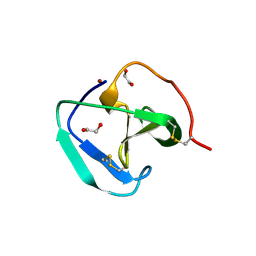 | | SH3-like domain from Penicillium virgatum muramidase | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2F
 
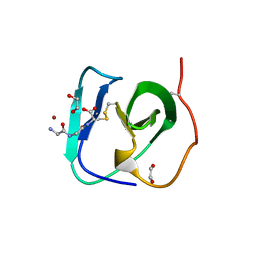 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | Descriptor: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2H
 
 | | Muramidase from Thermothielavioides terrestris, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2S
 
 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | Descriptor: | GH24 family muramidase, POTASSIUM ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
1SZP
 
 | | A Crystal Structure of the Rad51 Filament | | Descriptor: | DNA repair protein RAD51, SULFATE ION | | Authors: | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
