3M97
 
 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5IR6
 
 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2016-03-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
1W2L
 
 | | Cytochrome c domain of caa3 oxygen oxidoreductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CYTOCHROME OXIDASE SUBUNIT II, ... | | Authors: | Srinivasan, V, Rajendran, C, Sousa, F.L, Melo, A.M.P, Saraiva, L.M, Pereira, M.M, Santana, M, Teixeira, M, Michel, H. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure at 1.3 A resolution of Rhodothermus marinus caa(3) cytochrome c domain.
J. Mol. Biol., 345, 2005
|
|
1ZCD
 
 | | Crystal structure of the Na+/H+ antiporter NhaA | | Descriptor: | Na(+)/H(+) antiporter 1 | | Authors: | Hunte, C, Screpanti, E, Venturi, M, Rimon, A, Padan, E, Michel, H. | | Deposit date: | 2005-04-11 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH.
Nature, 435, 2005
|
|
1FNQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> GLU FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
1F6N
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> TYR FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-06-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
1FNP
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> PHE FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
1LGH
 
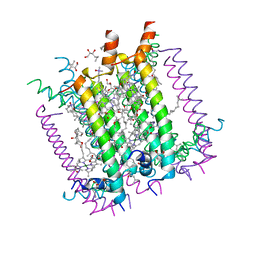 | | CRYSTAL STRUCTURE OF THE LIGHT-HARVESTING COMPLEX II (B800-850) FROM RHODOSPIRILLUM MOLISCHIANUM | | Descriptor: | BACTERIOCHLOROPHYLL A, HEPTANE-1,2,3-TRIOL, LIGHT HARVESTING COMPLEX II, ... | | Authors: | Koepke, J, Hu, X, Schulten, K, Michel, H. | | Deposit date: | 1996-03-20 | | Release date: | 1997-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum.
Structure, 4, 1996
|
|
7AU6
 
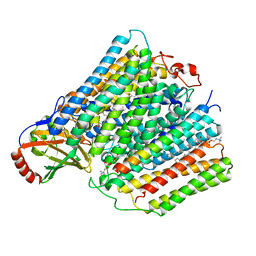 | |
7ATN
 
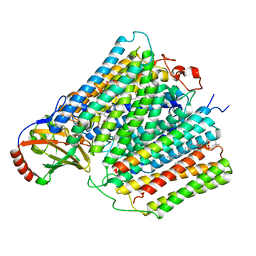 | |
7ATE
 
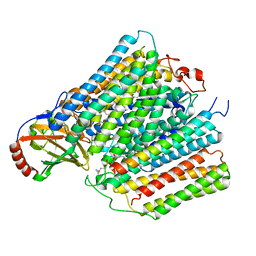 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
7AU3
 
 | | Cytochrome c oxidase structure in F-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cytochrome c oxidase structure in F-state
To Be Published
|
|
8B4O
 
 | | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome BD ubiquinol oxidase subunit I, Cytochrome bd-type quinol oxidase subunit II, ... | | Authors: | Grund, T.N, Kusumoto, T, Michel, H, Sakamoto, J, Safarian, S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum
To be published
|
|
3HYV
 
 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase from the hyperthermophilic bacterium Aquifex aeolicus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HYX
 
 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase from Aquifex aeolicus in complex with Aurachin C | | Descriptor: | 1-hydroxy-2-methyl-3-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]quinolin-4(1H)-one, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HYW
 
 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase of the hyperthermophilic bacterium Aquifex aeolicus in complex with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QL3
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the reduced state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QL4
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
6F27
 
 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6FV6
 
 | | Monomer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Monomer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV8
 
 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimer structure of the MATE family multidrug resistance transporter Aq128
in the outward-facing state
To Be Published
|
|
6HFB
 
 | | Outward-facing conformation of a multidrug resistance MATE family transporter of the MOP superfamily. | | Descriptor: | CESIUM ION, Uncharacterized protein | | Authors: | Nonaka, T, Zakrzewska, S, Safarian, S, Michel, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FV7
 
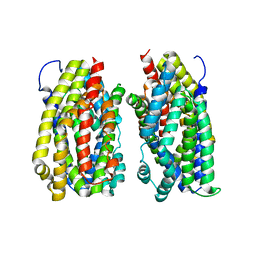 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dimer structure of Aq128 in the outward-facing state
To Be Published
|
|
