5E00
 
 | | Structure of HLA-A2 P130 | | 分子名称: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | 著者 | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | 登録日 | 2015-09-26 | | 公開日 | 2017-01-18 | | 最終更新日 | 2019-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
4WOY
 
 | | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1 | | 分子名称: | Mitochondrial dynamics protein MID49 | | 著者 | Loson, O.C, Meng, S, Ngo, H.B, Liu, R, Kaiser, J.T, Chan, D.C. | | 登録日 | 2014-10-17 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1.
Protein Sci., 24, 2015
|
|
6LKC
 
 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | 分子名称: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | 登録日 | 2019-12-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
5WSH
 
 | | Structure of HLA-A2 P130 | | 分子名称: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | 著者 | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | 登録日 | 2016-12-07 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-01-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
7ZJF
 
 | | R399E, a mutated form of GDF5, for disease modification of osteoarthritis | | 分子名称: | Growth/differentiation factor 5 | | 著者 | Gigout, A, Wekmann, D, Menges, S, Brenneis, C, Henson, F, Cowan, K.J, Musil, D, Thudium, C, Guehring, H, Michaelis, M, Kleinschmidt-Doerr, K. | | 登録日 | 2022-04-10 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | R399E, A Mutated Form of Growth and Differentiation Factor 5, for Disease Modification of Osteoarthritis.
Arthritis Rheumatol, 75, 2023
|
|
2IXZ
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | 分子名称: | 5'-R(*GP*CP*UP*GP*UP*GP*CP*CP)-3' | | 著者 | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-09-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2IXY
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | 分子名称: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | 著者 | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-09-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2OJ8
 
 | | NMR structure of the UGUU tetraloop of Duck Epsilon apical stem loop of the Hepatitis B virus | | 分子名称: | 5'-R(P*GP*CP*UP*GP*UP*UP*GP*U)-3' | | 著者 | Girard, F.C, Ottink, O.M, Ampt, K.A.M, Tessari, M, Wijmenga, S.S. | | 登録日 | 2007-01-12 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals.
Nucleic Acids Res., 35, 2007
|
|
2F1Q
 
 | |
1O8T
 
 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | 分子名称: | APOLIPOPROTEIN C-II | | 著者 | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | 登録日 | 2002-11-29 | | 公開日 | 2003-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
2K5Z
 
 | |
2OJ7
 
 | | NMR structure of the UGUU tetraloop of Duck Epsilon apical stem loop | | 分子名称: | 5'-R(P*GP*CP*UP*GP*UP*UP*GP*U)-3' | | 著者 | Girard, F.C, Ottink, O.M, Ampt, K.A.M, Tessari, M, Wijmenga, S.S. | | 登録日 | 2007-01-12 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals.
Nucleic Acids Res., 35, 2007
|
|
1AC7
 
 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | 著者 | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | 登録日 | 1997-02-14 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
1B4Y
 
 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | 分子名称: | DNA (H-Y5 TRIPLE HELIX) | | 著者 | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | 登録日 | 1998-12-30 | | 公開日 | 1999-09-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1A60
 
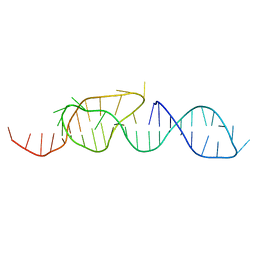 | | NMR STRUCTURE OF A CLASSICAL PSEUDOKNOT: INTERPLAY OF SINGLE-AND DOUBLE-STRANDED RNA, 24 STRUCTURES | | 分子名称: | TYMV PSEUDOKNOT | | 著者 | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | 登録日 | 1998-03-04 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA.
Science, 280, 1998
|
|
1EZN
 
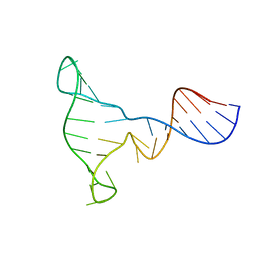 | | SOLUTION STRUCTURE OF A DNA THREE-WAY JUNCTION | | 分子名称: | DNA THREE-WAY JUNCTION | | 著者 | van Buuren, B.N.M, Overmars, F.J, Ippel, J.H, Altona, C, Wijmenga, S.S. | | 登録日 | 2000-05-11 | | 公開日 | 2001-04-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a DNA three-way junction containing two unpaired thymidine bases. Identification of sequence features that decide conformer selection.
J.Mol.Biol., 304, 2000
|
|
3PHP
 
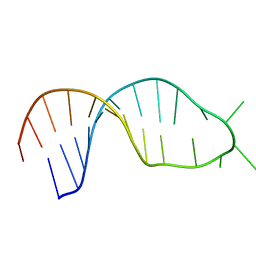 | | STRUCTURE OF THE 3' HAIRPIN OF THE TYMV PSEUDOKNOT: PREFORMATION IN RNA FOLDING | | 分子名称: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*GP*AP*GP*GP*GP*UP*CP*AP*UP*CP*GP*GP*AP*AP*CP*CP*A) -3') | | 著者 | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | 登録日 | 1998-10-13 | | 公開日 | 1998-10-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the 3'-hairpin of the TYMV pseudoknot: preformation in RNA folding.
EMBO J., 17, 1998
|
|
1BW5
 
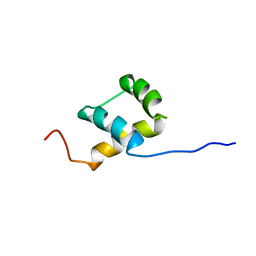 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | 分子名称: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | 著者 | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | 登録日 | 1998-09-29 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
1SNJ
 
 | | Solution structure of the DNA three-way junction with the A/C-stacked conformation | | 分子名称: | 36-MER | | 著者 | Wu, B, Girard, F, van Buuren, B, Schleucher, J, Tessari, M, Wijmenga, S. | | 登録日 | 2004-03-11 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold.
Nucleic Acids Res., 32, 2004
|
|
2JQ3
 
 | | Structure and Dynamics of Human Apolipoprotein C-III | | 分子名称: | Apolipoprotein C-III | | 著者 | Gangabadage, C.S, Zdunek, J, Tessari, M, Nilsson, S, Olivecrona, G, Wijmenga, S. | | 登録日 | 2007-05-28 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Dynamics of Human Apolipoprotein CIII
J.Biol.Chem., 283, 2008
|
|
