4HB1
 
 | | A DESIGNED FOUR HELIX BUNDLE PROTEIN. | | Descriptor: | DHP1, UNKNOWN ATOM OR ION | | Authors: | Schafmeister, C.E, Laporte, S.L, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 1997-11-10 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A designed four helix bundle protein with native-like structure.
Nat.Struct.Biol., 4, 1997
|
|
2JCB
 
 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3F66
 
 | | Human c-Met Kinase in complex with quinoxaline inhibitor | | Descriptor: | 3-[3-(4-methylpiperazin-1-yl)-7-(trifluoromethyl)quinoxalin-5-yl]phenol, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor, ... | | Authors: | Meier, C, Ceska, T. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a novel series of quinoxalines as inhibitors of c-Met kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2CME
 
 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
2JJX
 
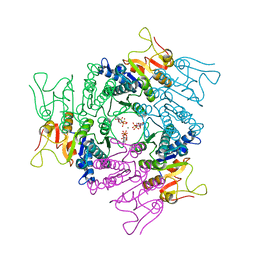 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
1AR1
 
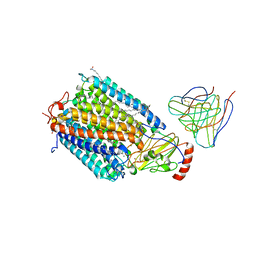 | | Structure at 2.7 Angstrom Resolution of the Paracoccus Denitrificans two-subunit Cytochrome C Oxidase Complexed with an Antibody Fv Fragment | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ostermeier, C, Harrenga, A, Ermler, U, Michel, H. | | Deposit date: | 1997-08-08 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
3SLS
 
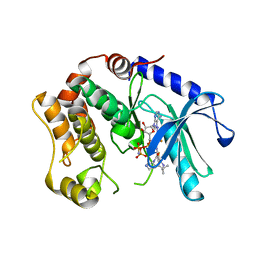 | | Crystal Structure of human MEK-1 kinase in complex with UCB1353770 and AMPPNP | | Descriptor: | 2-[(2-fluoro-4-iodophenyl)amino]-5,5-dimethyl-8-oxo-N-[(3R)-piperidin-3-yl]-5,6,7,8-tetrahydro-4H-thieno[2,3-c]azepine-3-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Meier, C, Ceska, T.A. | | Deposit date: | 2011-06-25 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting.
J.Struct.Biol., 177, 2012
|
|
2I2X
 
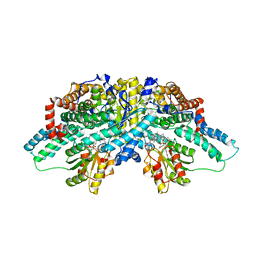 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZBD
 
 | |
1QV9
 
 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
6A70
 
 | | Structure of the human PKD1/PKD2 complex | | Descriptor: | Polycystin-1, Polycystin-2 | | Authors: | Su, Q, Hu, F, Ge, X, Lei, J, Yu, S, Wang, T, Zhou, Q, Mei, C, Shi, Y. | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human PKD1-PKD2 complex.
Science, 361, 2018
|
|
8RFK
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RE0
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
4BLQ
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI8 | | Descriptor: | P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BLO
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI6 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, PACKAGING ENZYME P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
5MIN
 
 | | Apo form of the soluble PQQ-dependent Glucose Dehydrogenase from Acinetobacter calcoaceticus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Quinoprotein glucose dehydrogenase B | | Authors: | Stines-Chaumeil, C, Mavre, F, Limoges, B, Kauffmann, B, Mano, N. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Apo form of the soluble PQQ-dependent Glucose Dehydrogenase from Acinetobacter calcoaceticus
To Be Published
|
|
4FGI
 
 | | Structure of the effector - immunity system Tse1 / Tsi1 from Pseudomonas aeruginosa | | Descriptor: | Tse1, Tsi1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FGE
 
 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, THIOCYANATE ION, ... | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FGD
 
 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa, selenomethionine variant | | Descriptor: | CHLORIDE ION, THIOCYANATE ION, Tse1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
7BBR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCP
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with gluconate | | Descriptor: | D-gluconic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCO
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with D-foconate | | Descriptor: | (2R,3S,4S,5S)-2,3,4,5-tetrahydroxyhexanoic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with galactonate | | Descriptor: | L-galactonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCN
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with Xylonic acid | | Descriptor: | D-xylonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
