1M5Y
 
 | |
1HMH
 
 | |
1G41
 
 | | CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1BA1
 
 | |
1BUP
 
 | | T13S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sousa, M.C, Mckay, D.B. | | Deposit date: | 1998-09-03 | | Release date: | 1998-09-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
2AVU
 
 | | Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription | | Descriptor: | Flagellar transcriptional activator flhC, Transcriptional activator flhD, ZINC ION | | Authors: | Wang, S, Fleming, R.T, Westbrook, E.M, Matsumura, P, McKay, D.B. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Escherichia coli FlhDC Complex, a Prokaryotic Heteromeric Regulator of Transcription.
J.Mol.Biol., 355, 2006
|
|
1BA0
 
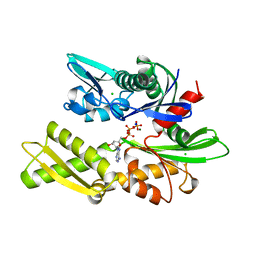 | | HEAT-SHOCK COGNATE 70KD PROTEIN 44KD ATPASE N-TERMINAL 1NGE 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HEAT-SHOCK COGNATE 70KD PROTEIN, ... | | Authors: | Wilbanks, S.M, Mckay, D.B. | | Deposit date: | 1998-04-21 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural replacement of active site monovalent cations by the epsilon-amino group of lysine in the ATPase fragment of bovine Hsc70.
Biochemistry, 37, 1998
|
|
1EZM
 
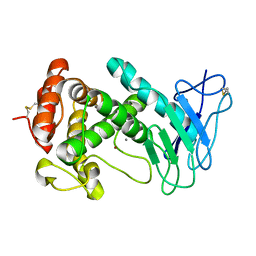 | |
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G3K
 
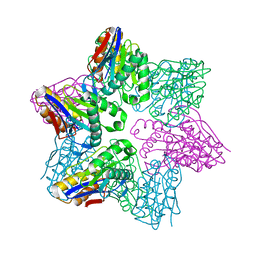 | |
1OFI
 
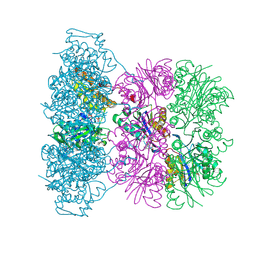 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1FUU
 
 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FUK
 
 | |
1QQM
 
 | | D199S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D199S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN, ... | | Authors: | Johnson, E.R, Mckay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1QQO
 
 | | E175S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HYDROLASE (ACTING ON ACID ANHYDRIDES), ... | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1QQN
 
 | | D206S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D206S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN, ... | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1NGJ
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGD
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGH
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGC
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGI
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | CALCIUM ION, HEAT-SHOCK COGNATE 70 kD PROTEIN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGA
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGG
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGB
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
