3ONS
 
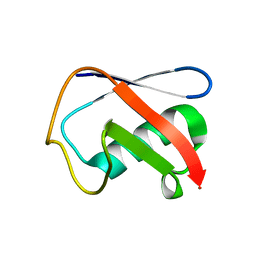 | | Crystal structure of Human Ubiquitin in a new crystal form | | 分子名称: | Ubiquitin | | 著者 | Amodeo, G.A, Huang, K.Y, Mcdermott, A.E, Tong, L. | | 登録日 | 2010-08-30 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of human ubiquitin in 2-methyl-2,4-pentanediol: a new conformational switch.
Protein Sci., 20, 2011
|
|
1I45
 
 | |
4KPB
 
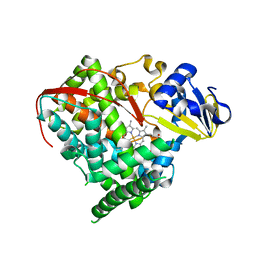 | | Crystal structure of cytochrome P450 BM-3 R47E mutant | | 分子名称: | Cytochrome P450 BM-3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sadre-Bazzaz, K, Catalano, J, McDermott, A.E, Tong, L. | | 登録日 | 2013-05-13 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Evidence: A Single Charged Residue Affects Substrate Binding in Cytochrome P450 BM-3.
Biochemistry, 52, 2013
|
|
4KPA
 
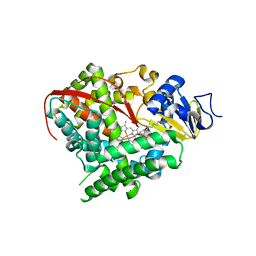 | | Crystal structure of cytochrome P450 BM-3 in complex with N-palmitoylglycine (NPG) | | 分子名称: | Cytochrome P450 BM-3, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Amadeo, G.A, Catalano, J, McDermott, A.E, Tong, L. | | 登録日 | 2013-05-13 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Evidence: A Single Charged Residue Affects Substrate Binding in Cytochrome P450 BM-3.
Biochemistry, 52, 2013
|
|
1NEY
 
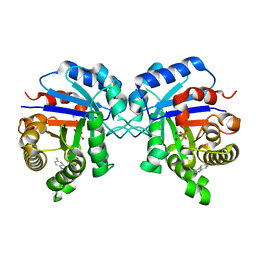 | | Triosephosphate Isomerase in Complex with DHAP | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | 著者 | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | 登録日 | 2002-12-12 | | 公開日 | 2003-01-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NF0
 
 | | Triosephosphate Isomerase in Complex with DHAP | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | 著者 | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | 登録日 | 2002-12-12 | | 公開日 | 2003-01-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5V7Z
 
 | | SSNMR Structure of the Human RIP1/RIP3 Necrosome | | 分子名称: | PRO-LEU-VAL-ASN-ILE-TYR-ASN-CYS-SER-GLY-VAL-GLN-VAL-GLY-ASP, THR-ILE-TYR-ASN-SER-THR-GLY-ILE-GLN-ILE-GLY-ALA-TYR-ASN-TYR-MET-GLU-ILE | | 著者 | Mompean, M, Li, W, Li, J, Laage, S, Siemer, A.B, Wu, H, McDermott, A.E. | | 登録日 | 2017-03-21 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex.
Cell, 173, 2018
|
|
