3PNL
 
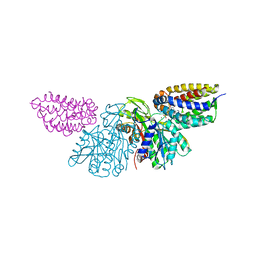 | | Crystal Structure of E.coli Dha kinase DhaK-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNO
 
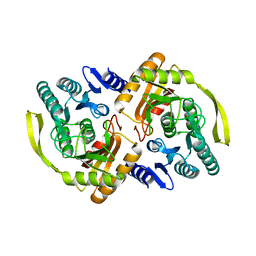 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) | | Descriptor: | PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNQ
 
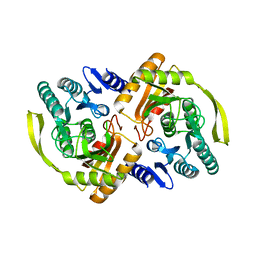 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNM
 
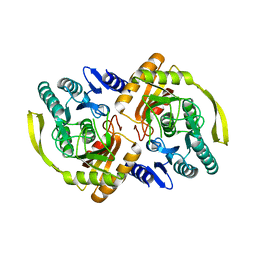 | | Crystal Structure of E.coli Dha kinase DhaK (H56A) | | Descriptor: | PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNK
 
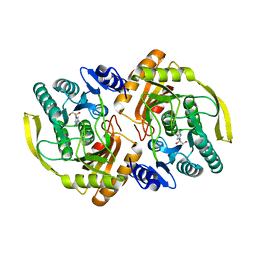 | | Crystal Structure of E.coli Dha kinase DhaK | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2OV8
 
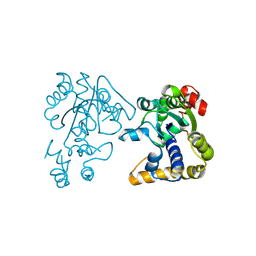 | |
3B8N
 
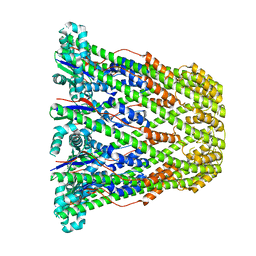 | |
3B8O
 
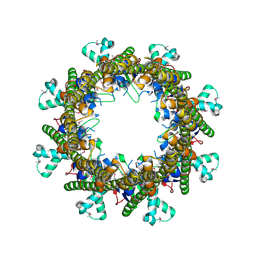 | |
3B8P
 
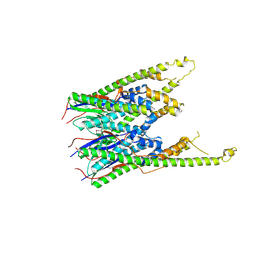 | |
3LA0
 
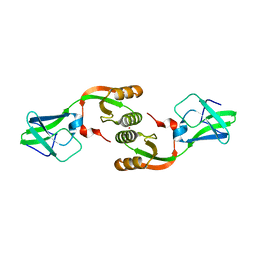 | | Crystal Structure of UreE from Helicobacter pylori (metal of unknown identity bound) | | Descriptor: | UNKNOWN ATOM OR ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3L9Z
 
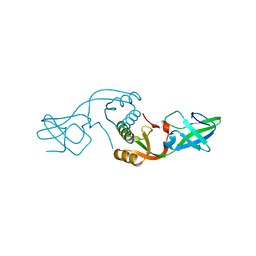 | | Crystal Structure of UreE from Helicobacter pylori (apo form) | | Descriptor: | Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3NY0
 
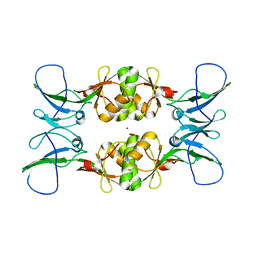 | | Crystal Structure of UreE from Helicobacter pylori (Ni2+ bound form) | | Descriptor: | NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3NXZ
 
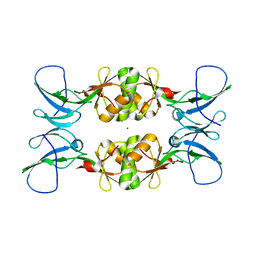 | | Crystal Structure of UreE from Helicobacter pylori (Cu2+ bound form) | | Descriptor: | COPPER (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3BE5
 
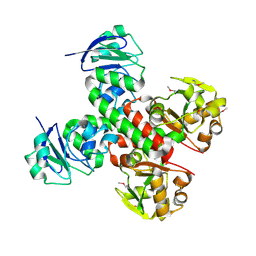 | | Crystal structure of FitE (crystal form 1), a group III periplasmic siderophore binding protein | | Descriptor: | CHLORIDE ION, Putative iron compound-binding protein of ABC transporter family | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-16 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trapping open and closed forms of FitE-A group III periplasmic binding protein.
Proteins, 75, 2008
|
|
1SBZ
 
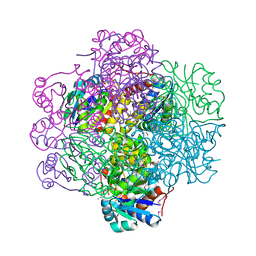 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
3LVL
 
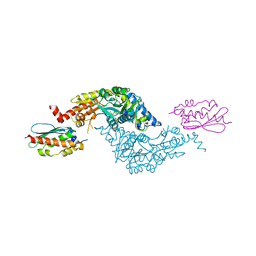 | | Crystal Structure of E.coli IscS-IscU complex | | Descriptor: | Cysteine desulfurase, NifU-like protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVJ
 
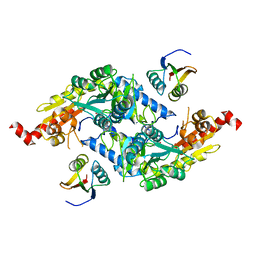 | | Crystal Structure of E.coli IscS-TusA complex (form 1) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVK
 
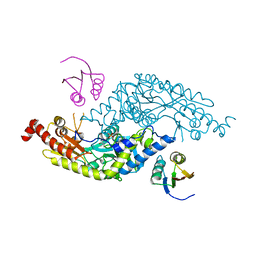 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVM
 
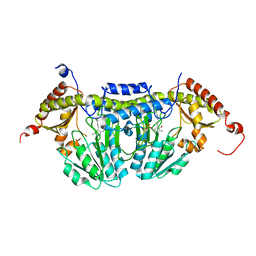 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3CES
 
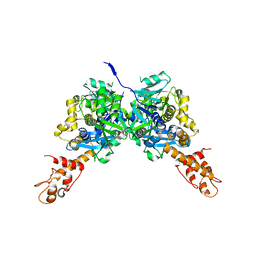 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3G05
 
 | | Crystal structure of N-terminal domain (2-550) of E.coli MnmG | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
1FC4
 
 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1SZ2
 
 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
