6YMZ
 
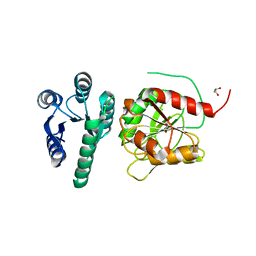 | | Structure of the CheB methylsterase from P. atrosepticum SCRI1043 | | Descriptor: | ACETATE ION, GLYCEROL, Protein-glutamate methylesterase/protein-glutamine glutaminase, ... | | Authors: | Gavira, J.A, Krell, T, Velando-Soriano, F, Matilla, M.A. | | Deposit date: | 2020-04-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Pentapeptide-Dependent and Independent CheB Methylesterases.
Int J Mol Sci, 21, 2020
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7QEJ
 
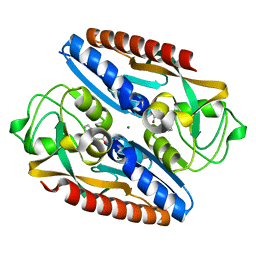 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7PSG
 
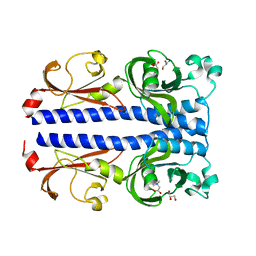 | | Structure of the ligand binding domain of the PacA (ECA2226) chemoreceptor of Pectobacterium atrosepticum SCRI1043 in complex with betaine. | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, TRIMETHYL GLYCINE | | Authors: | Gavira, J.A, Matilla, M.A, Velando, F, Krell, T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRR
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRQ
 
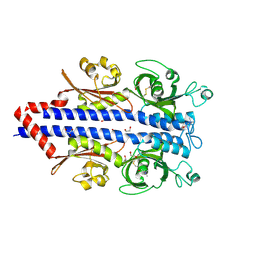 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with choline. | | Descriptor: | 1,2-ETHANEDIOL, CHOLINE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
6FU4
 
 | | Ligand binding domain (LBD) of the p. aeruginosa histamine receptor TlpQ | | Descriptor: | ACETATE ION, GLYCEROL, HISTAMINE, ... | | Authors: | Gavira, J.A, Krell, T, Conejero-Muriel, M, Corral-Lugo, A, Matilla, M.A, Silva Jimenez, H, Mesa Torres, N, Martin-Mora, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | High-Affinity Chemotaxis to Histamine Mediated by the TlpQ Chemoreceptor of the Human Pathogen Pseudomonas aeruginosa.
MBio, 9, 2018
|
|
6S1A
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP | | Descriptor: | Aromatic acid chemoreceptor, SULFATE ION | | Authors: | Gavira, J.A, Matilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
