2MWD
 
 | |
6ZMU
 
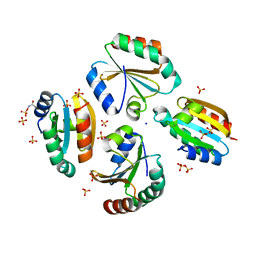 | | Crystal structure of the germline-specific thioredoxin protein Deadhead (Thioredoxin-1) from Drospohila melanogaster, P43212 | | Descriptor: | SODIUM ION, SULFATE ION, Thioredoxin-1 | | Authors: | Baginski, B, Pluta, R, Macias, M.J. | | Deposit date: | 2020-07-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
6ZMN
 
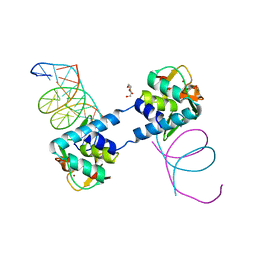 | |
7PVU
 
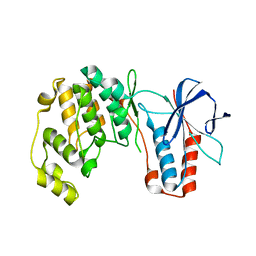 | | Crystal structure of p38alpha C162S in complex with CAS2094511-69-8, P 1 21 1 | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)-2-fluorobenzene-1-sulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2021-10-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
2B7E
 
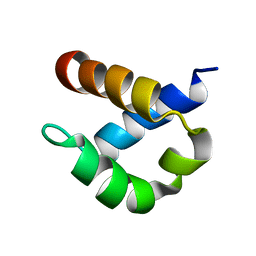 | | First FF domain of Prp40 Yeast Protein | | Descriptor: | Pre-mRNA processing protein PRP40 | | Authors: | Gasch, A, Wiesner, S, Martin-Malpartida, P, Ramirez-Espain, X, Ruiz, L, Macias, M.J. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains.
J.Biol.Chem., 281, 2006
|
|
1MPH
 
 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
8ACO
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACM
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
2RM0
 
 | | FBP28WW2 domain in complex with a PPPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2RLY
 
 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
1W4M
 
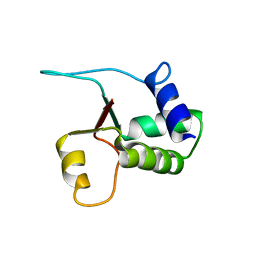 | | Structure of the human pleckstrin DEP domain by multidimensional NMR | | Descriptor: | PLECKSTRIN | | Authors: | Civera, C, Simon, B, Stier, G, Sattler, M, Macias, M.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Human Pleckstrin Dep Domain: Distinct Molecular Features of a Novel Dep Domain Subfamily
Proteins: Struct.,Funct., Genet., 58, 2005
|
|
1YIU
 
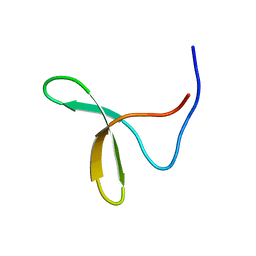 | | Itch E3 ubiquitin ligase WW3 domain | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Shaw, A.Z, Martin-Malpartida, P, Morales, B, Yraola, F, Royo, M, Macias, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of either Ser16 or Thr30 does not disrupt the structure of the Itch E3 ubiquitin ligase third WW domain
Proteins, 60, 2005
|
|
6TBZ
 
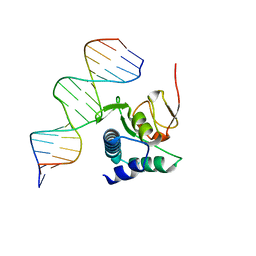 | |
6TCE
 
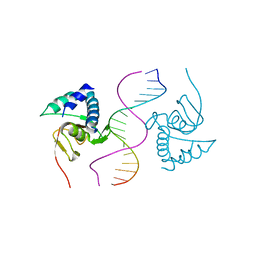 | |
1O6W
 
 | |
1JMQ
 
 | | YAP65 (L30K mutant) WW domain in Complex with GTPPPPYTVG peptide | | Descriptor: | 65 KDA YES-ASSOCIATED PROTEIN, WW Domain Binding Protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-07-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1K9Q
 
 | | YAP65 WW domain complexed to N-(n-octyl)-GPPPY-NH2 | | Descriptor: | 65 kDa Yes-associated protein, N-OCTANE, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1K9R
 
 | | YAP65 WW domain complexed to Acetyl-PLPPY | | Descriptor: | 65 kDa Yes-associated protein, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1D8B
 
 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
7YZA
 
 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGTATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZ7
 
 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZB
 
 | | Crystal structure of the human FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZD
 
 | |
7YZC
 
 | | Crystal structure of the zebrafish FoxH1 bound to the TGTTTATT site | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*TP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*AP*AP*CP*AP*AP*TP*CP*T)-3'), ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZG
 
 | | Crystal structure of the Xenopus FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*TP*G)-3'), Forkhead box protein H1 | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
