3ZK0
 
 | |
3CFZ
 
 | |
3ZP5
 
 | | Crystal structure of the DNA binding ETS domain of the human protein FEV in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', PROTEIN FEV | | Authors: | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Allerston, C.K, Vollmar, M, Arrowsmith, C.H, Burgess-Brown, N, Edwards, A, von Delft, F, Gileadi, O. | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4ONW
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ACETATE ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4OXI
 
 | | Crystal structure of Vibrio cholerae adenylation domain AlmE in complex with glycyl-adenosine-5'-phosphate | | Descriptor: | Enterobactin synthetase component F-related protein, GLYCYL-ADENOSINE-5'-PHOSPHATE | | Authors: | Fage, C.D, Henderson, J.C, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases.
Acs Chem.Biol., 9, 2014
|
|
3CFX
 
 | | Crystal structure of M. acetivorans periplasmic binding protein ModA/WtpA with bound tungstate | | Descriptor: | GLYCEROL, MAGNESIUM ION, TUNGSTATE(VI)ION, ... | | Authors: | Comellas-Bigler, M, Hollenstein, K, Locher, K.P. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distorted octahedral coordination of tungstate in a subfamily of specific binding proteins.
J.Biol.Inorg.Chem., 14, 2009
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
3D4U
 
 | | Bovine thrombin-activatable fibrinolysis inhibitor (TAFIa) in complex with tick-derived carboxypeptidase inhibitor. | | Descriptor: | ACETATE ION, Carboxypeptidase B2, Carboxypeptidase inhibitor, ... | | Authors: | Sanglas, L, Valnickova, Z, Arolas, J.L, Pallares, I, Guevara, T, Sola, M, Kristensen, T, Enghild, J.J, Aviles, F.X, Gomis-Ruth, F.X. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of activated thrombin-activatable fibrinolysis inhibitor, a molecular link between coagulation and fibrinolysis.
Mol.Cell, 31, 2008
|
|
4P9X
 
 | | Structure of ConA/Rh3Glu complex | | Descriptor: | 2-[2-(2-{4-[(alpha-D-glucopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Sakai, F, Weiss, M.S, Chen, G. | | Deposit date: | 2014-04-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of ConA/Rh3Glu complex
To Be Published
|
|
8VI2
 
 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VKD
 
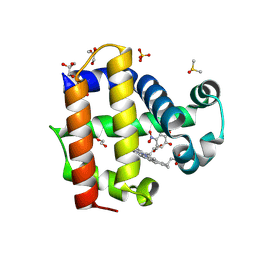 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
4P1M
 
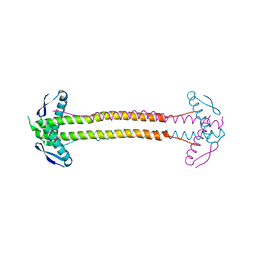 | | The structure of Escherichia coli ZapA | | Descriptor: | CHLORIDE ION, Cell division protein ZapA | | Authors: | Kimber, M.S, Roach, E.J, Khursigara, C.M. | | Deposit date: | 2014-02-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-directed Mutational Analysis Reveals Key Residues Involved in Escherichia coli ZapA Function.
J.Biol.Chem., 289, 2014
|
|
4PAC
 
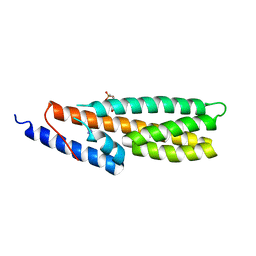 | | Crystal Structure of Histidine-containing Phosphotransfer Protein AHP2 from Arabidopsis thaliana | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Histidine-containing phosphotransfer protein 2, IMIDAZOLE | | Authors: | Degtjarik, O, Dopitova, R, Puehringer, S, Weiss, M.S, Janda, L, Hejatko, J, Kuta-Smatanova, I. | | Deposit date: | 2014-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of AHP2 from Arabidopsis thaliana
To Be Published
|
|
8XEQ
 
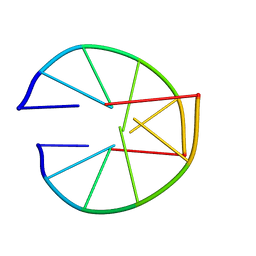 | |
8XGW
 
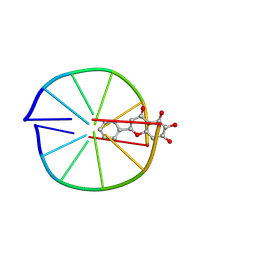 | |
4BBO
 
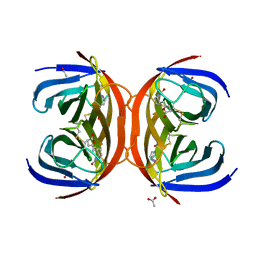 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
3CQL
 
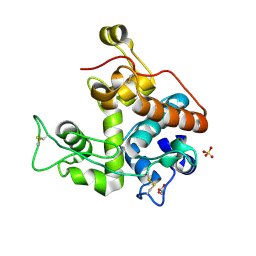 | | Crystal Structure of GH family 19 chitinase from Carica papaya | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endochitinase, ... | | Authors: | Huet, J, Rucktoa, P, Clantin, B, Azarkan, M, Looze, Y, Villeret, V, Wintjens, R. | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray Structure of Papaya Chitinase Reveals the Substrate Binding Mode of Glycosyl Hydrolase Family 19 Chitinases.
Biochemistry, 47, 2008
|
|
8VZR
 
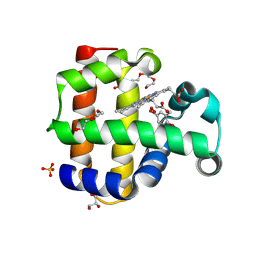 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VSK
 
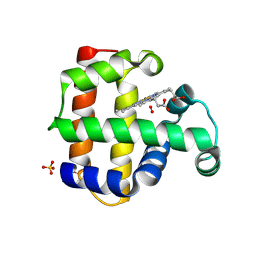 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VWR
 
 | | Structure of steroid hydratase from Comamonas testosteroni | | Descriptor: | Acyl dehydratase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schroeter, K.L, Forrester, T.J.B, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Shy is a proteobacterial steroid hydratase which catalyzes steroid side chain degradation without requiring a catalytically inert partner domain.
J.Biol.Chem., 300, 2024
|
|
4NOA
 
 | |
8YA8
 
 | |
4O1E
 
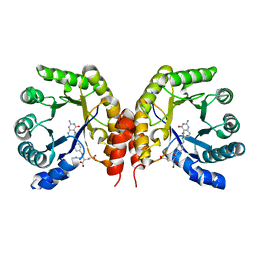 | | Structure of a methyltransferase component in complex with MTHF involved in O-demethylation | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P9Y
 
 | | Structure of ConA/Rh4man | | Descriptor: | 2-{2-[2-(2-{4-[(alpha-D-mannopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethoxy}ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Sakai, F, Weiss, M.S, Chen, G. | | Deposit date: | 2014-04-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of ConA/Rh4Man
To Be Published
|
|
3ATJ
 
 | | HEME LIGAND MUTANT OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (HORSERADISH PEROXIDASE C1A), ... | | Authors: | Meno, K, White, C.G, Smith, A.T, Gajhede, M. | | Deposit date: | 1998-12-16 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Catalytical Implications of a F221M Mutation in the Proximal Pocket of Horseradish Peroxidase C (HRP C)
To be Published
|
|
