6ZF4
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(2,3,5,6-tetramethylphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
1QHD
 
 | | CRYSTAL STRUCTURE OF VP6, THE MAJOR CAPSID PROTEIN OF GROUP A ROTAVIRUS | | Descriptor: | CALCIUM ION, CHLORIDE ION, VIRAL CAPSID VP6, ... | | Authors: | Mathieu, M, Petitpas, I, Rey, F.A. | | Deposit date: | 1999-04-29 | | Release date: | 2001-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion.
EMBO J., 20, 2001
|
|
6ZF7
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6Z4R
 
 | |
6Z4T
 
 | |
6ESF
 
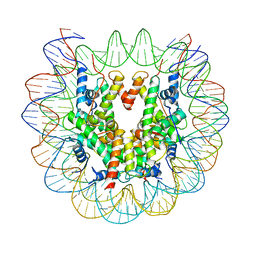 | | Nucleosome : Class 1 | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7QW8
 
 | | Adenine-specific DNA methyltransferase M.BseCI | | Descriptor: | Modification methylase BseCI | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QW7
 
 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate fully methylated DNA duplex | | Descriptor: | Fully methylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QQ7
 
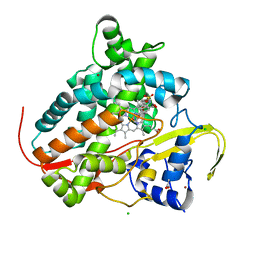 | |
7QW5
 
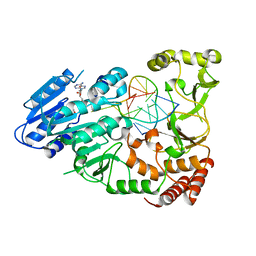 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate unmethylated DNA duplex | | Descriptor: | Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE, Unmethylated DNA duplex | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7LD9
 
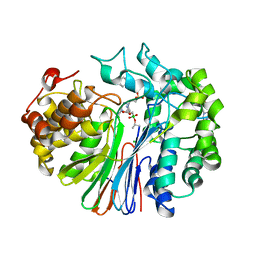 | | Structure of human GGT1 in complex with ABBA | | Descriptor: | (2R)-2-amino-4-boronobutanoic acid, (2S)-2-amino-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
6EJN
 
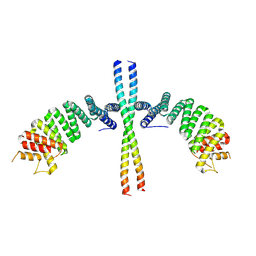 | |
7QW6
 
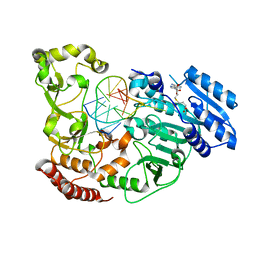 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate hemimethylated DNA duplex | | Descriptor: | Hemimethylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
6YZH
 
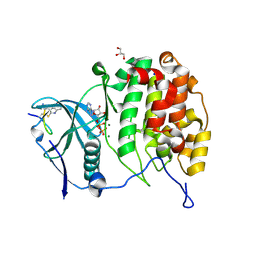 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
7QXF
 
 | |
1QAC
 
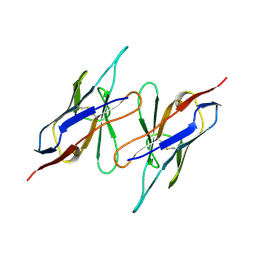 | | CHANGE IN DIMERIZATION MODE BY REMOVAL OF A SINGLE UNSATISFIED POLAR RESIDUE | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN VARIABLE DOMAIN | | Authors: | Pokkuluri, P.R, Cai, X, Johnson, G, Stevens, F.J, Schiffer, M. | | Deposit date: | 1999-02-25 | | Release date: | 2000-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Change in dimerization mode by removal of a single unsatisfied polar residue located at the interface.
Protein Sci., 9, 2000
|
|
4MML
 
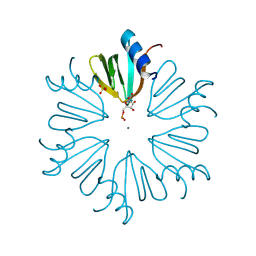 | | D40A Hfq from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Protein hfq, SULFATE ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
6EE0
 
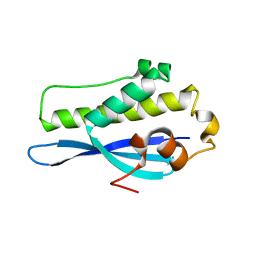 | | Crystal Structure of SNX23 PX domain | | Descriptor: | Kinesin-like protein KIF16B | | Authors: | Chandra, M, Collins, B.M. | | Deposit date: | 2018-08-12 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities.
Nat Commun, 10, 2019
|
|
1QD6
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CALCIUM ION, OUTER MEMBRANE PHOSPHOLIPASE (OMPLA), ... | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
6Z19
 
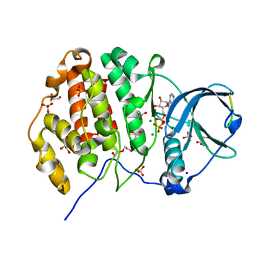 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6EM9
 
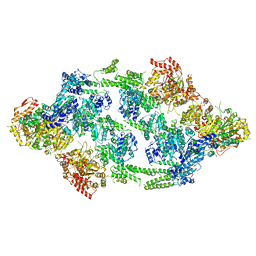 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
1QVO
 
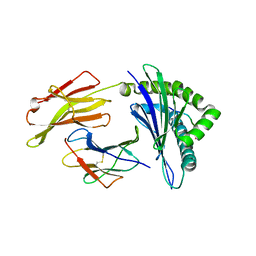 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
6Z98
 
 | | NMR solution structure of the peach allergen Pru p 1.0101 | | Descriptor: | Major allergen Pru p 1 | | Authors: | Eidelpes, R, Fuehrer, S, Hofer, F, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Zeatin Binding of the Peach Allergen Pru p 1 .
J.Agric.Food Chem., 69, 2021
|
|
3ZUK
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE ZMP1 IN COMPLEX WITH INHIBITOR | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ferraris, D.M, Sbardella, D, Petrera, A, Marini, S, Amstutz, B, Coletta, M, Sander, P, Rizzi, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Zinc-Dependent Metalloprotease-1 (Zmp1), a Metalloprotease Involved in Pathogenicity.
J.Biol.Chem., 286, 2011
|
|
6ZBK
 
 | | Crystal structure of the human complex between RPAP3 and TRBP | | Descriptor: | RISC-loading complex subunit TARBP2, RNA polymerase II-associated protein 3 | | Authors: | Charron, C, Abel, Y, Charpentier, B, Rederstorff, M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Nucleic Acids Res., 50, 2022
|
|
