5V0F
 
 | | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone | | 分子名称: | 4-arsanyl-2-nitrophenol, CHLORIDE ION, FE (III) ION, ... | | 著者 | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | 登録日 | 2017-02-28 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone.
To Be Published
|
|
5V45
 
 | |
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | 分子名称: | GLYCEROL, Putative translational inhibitor protein | | 著者 | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-03-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
2OYI
 
 | | Crystal Structure of Fragment D of gammaD298,301A Fibrinogen with the Peptide Ligand Gly-Pro-Arg-Pro-Amide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha chain, ... | | 著者 | Kostelansky, M.S, Gorkun, O.V, Lord, S.T. | | 登録日 | 2007-02-22 | | 公開日 | 2007-05-15 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Probing the gamma2 Calcium-Binding Site: Studies with gammaD298,301A Fibrinogen Reveal Changes in the gamma294-301 Loop that Alter the Integrity of the "a" Polymerization Site.
Biochemistry, 46, 2007
|
|
5UZR
 
 | |
6D8C
 
 | | Cryo-EM structure of FLNaABD E254K bound to phalloidin-stabilized F-actin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Iwamoto, D.V, Huehn, A.R, Simon, B, Huet-Calderwood, C, Baldassarre, M, Sindelar, C.V, Calderwood, D.A. | | 登録日 | 2018-04-26 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural basis of the filamin A actin-binding domain interaction with F-actin.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DA1
 
 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-01 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.000127 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
2P1I
 
 | | Plasmodium yoelii Ribonucleotide Reductase Subunit R2 (PY03671) | | 分子名称: | FE (III) ION, Ribonucleotide reductase, small chain | | 著者 | Wernimont, A.K, Dong, A, Choe, J, Gao, M, Walker, J, Lew, J, Alam, Z, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | 登録日 | 2007-03-05 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Plasmodium yoelii Ribonucleotide Reductase Subunit R2 (PY03671)
To be Published
|
|
5V2Z
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoadipic acid and L-arginine | | 分子名称: | 2-OXOADIPIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, ... | | 著者 | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | 登録日 | 2017-03-06 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | 分子名称: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5V46
 
 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | 分子名称: | Sacsin | | 著者 | Menade, M, Kozlov, G, Gehring, K. | | 登録日 | 2017-03-08 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
2P30
 
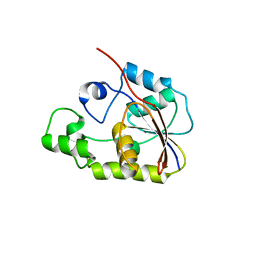 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | 分子名称: | Alpha-ribazole-5'-phosphate phosphatase | | 著者 | Sugahara, M, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-08 | | 公開日 | 2007-09-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
5UR0
 
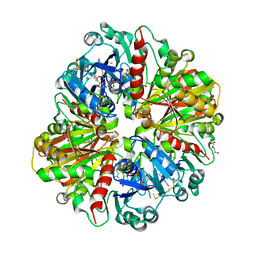 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | 著者 | Machado, A.T.P, Silva, M, Iulek, J. | | 登録日 | 2017-02-09 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural studies of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa.
Biochim. Biophys. Acta, 1866, 2018
|
|
2PCA
 
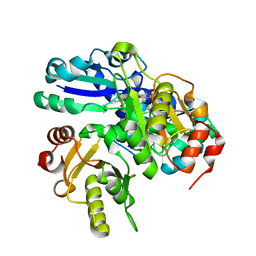 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | 分子名称: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | 著者 | Sugahara, M, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-29 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
3U8G
 
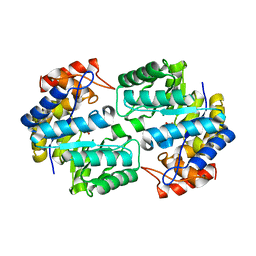 | | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with Oxalic acid at 1.80 A resolution | | 分子名称: | Dihydrodipicolinate synthase, OXALATE ION | | 著者 | Kumar, M, Kaushik, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2011-10-17 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with Oxalic acid at 1.80 A resolution
To be Published
|
|
5USW
 
 | | The crystal structure of 7,8-dihydropteroate synthase from Vibrio fischeri ES114 | | 分子名称: | ACETATE ION, Dihydropteroate synthase, FORMIC ACID, ... | | 著者 | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-14 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.643 Å) | | 主引用文献 | The crystal structure of 7,8-dihydropteroate synthase from Vibrio fischeri ES114
To Be Published
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.001 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5UVG
 
 | | Crystal structure of the human neutral sphingomyelinase 2 (nSMase2) catalytic domain with insertion deleted and calcium bound | | 分子名称: | CALCIUM ION, Sphingomyelin phosphodiesterase 3,Sphingomyelin phosphodiesterase 3 | | 著者 | Airola, M.V, Guja, K.E, Garcia-Diaz, M, Hannun, Y.A. | | 登録日 | 2017-02-20 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Structure of human nSMase2 reveals an interdomain allosteric activation mechanism for ceramide generation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6D4M
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG3 | | 著者 | Gohain, N, Tolbert, W.D, Pazgier, M. | | 登録日 | 2018-04-18 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D92
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-A non-canonical pair at position 3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-27 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
3WEL
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltotriose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, GLYCEROL, ... | | 著者 | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | 登録日 | 2013-07-08 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
2P0C
 
 | | Catalytic Domain of the Proto-oncogene Tyrosine-protein Kinase MER | | 分子名称: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-02-28 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
6H5S
 
 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | 分子名称: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | 著者 | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | 登録日 | 2018-07-25 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2P1Q
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | 登録日 | 2007-03-06 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Mechanism of auxin perception by the TIR1 ubiquitin ligase.
Nature, 446, 2007
|
|
