8AIA
 
 | |
8AIX
 
 | |
8AFE
 
 | |
8AFM
 
 | |
6F95
 
 | |
5F0N
 
 | | Cohesin subunit Pds5 | | Descriptor: | cohesin subunit Pds5,cohesin subunit Pds5, cohesin subunit Pds5,KLTH0D07062p,cohesin subunit Pds5,cohesin subunit Pds5, cohesin subunit Pds5 | | Authors: | Lee, B.-G, Jansma, M, Nasmyth, K, Lowe, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1.
Cell Rep, 14, 2016
|
|
5MW1
 
 | |
5DCF
 
 | | C-terminal domain of XerD recombinase in complex with gamma domain of FtsK | | Descriptor: | Tyrosine recombinase XerD,DNA translocase FtsK | | Authors: | Keller, A.N, Xin, Y, Lowe, J, Grainge, I. | | Deposit date: | 2015-08-24 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation of Xer-recombination at dif: structural basis of the FtsK gamma-XerD interaction.
Sci Rep, 6, 2016
|
|
4V03
 
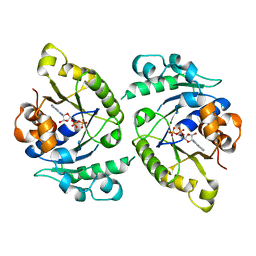 | | MinD cell division protein, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SITE-DETERMINING PROTEIN | | Authors: | Trambaiolo, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
5AI7
 
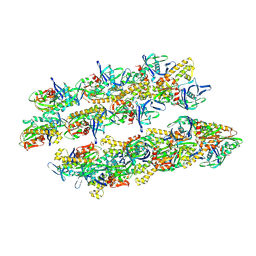 | | ParM doublet model | | Descriptor: | PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles
Nature, 523, 2015
|
|
5AEY
 
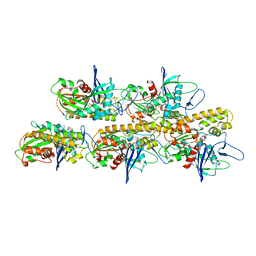 | | actin-like ParM protein bound to AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles.
Nature, 523, 2015
|
|
5N97
 
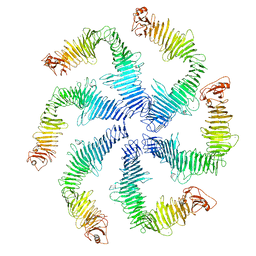 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
5FO3
 
 | | ZapC cell division regulator from E. coli | | Descriptor: | CELL DIVISION PROTEIN ZAPC | | Authors: | Ortiz, C, Kureisaite-Ciziene, D, Schmitz, F, Vicente, M, Lowe, J. | | Deposit date: | 2015-11-18 | | Release date: | 2015-11-25 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Z-Ring Associated Cell Division Protein Zapc from Escherichia Coli.
FEBS Lett., 589, 2015
|
|
8AFL
 
 | |
5LY5
 
 | | Arcadin-1 from Pyrobaculum calidifontis | | Descriptor: | arcadin-1 | | Authors: | Izore, T, Lowe, J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crenactin forms actin-like double helical filaments regulated by arcadin-2.
Elife, 5, 2016
|
|
5LY3
 
 | |
5NR1
 
 | | FzlA from C. crescentus | | Descriptor: | FtsZ-binding protein FzlA | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division.
Mol. Microbiol., 107, 2018
|
|
5O09
 
 | | BtubABC mini microtubule | | Descriptor: | Bacterial kinesin light chain, GUANOSINE-5'-DIPHOSPHATE, Tubulin, ... | | Authors: | Deng, X, Bharat, T.A.M, Lowe, J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4A6J
 
 | | Structural model of ParM filament based on CryoEM map | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Gayathri, P, Fujii, T, Moller-Jensen, J, Van Den Ent, F, Namba, K, Lowe, J. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
3M6A
 
 | |
3M65
 
 | |
4A61
 
 | | ParM from plasmid R1 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | van den Ent, F, Moller-Jensen, J, Gayathri, P, Lowe, J. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
4A62
 
 | |
3ZID
 
 | |
4A2A
 
 | | Thermotoga maritima FtsA:FtsZ(336-351) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, PUTATIVE, ... | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2011-09-23 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ftsa Forms Actin-Like Protofilaments
Embo J., 31, 2012
|
|
