6N88
 
 | |
6N89
 
 | |
6ESF
 
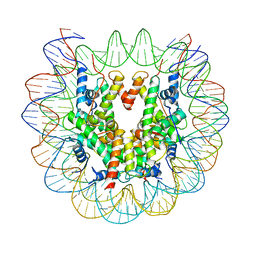 | | Nucleosome : Class 1 | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2017-10-20 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ESG
 
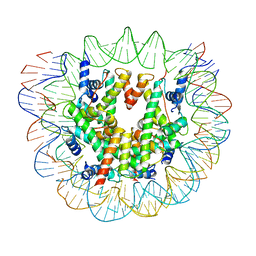 | | Nucleosome breathing : Class 2 | | 分子名称: | DNA (141-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2017-10-20 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ESH
 
 | | Nucleosome breathing : Class 3 | | 分子名称: | DNA (137-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2017-10-20 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ESI
 
 | | Nucleosome breathing : Class 4 | | 分子名称: | DNA (133-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2017-10-20 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FQ5
 
 | | Class 1 : canonical nucleosome | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2018-02-13 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural rearrangements of the histone octamer translocate DNA.
Nat Commun, 9, 2018
|
|
6FQ6
 
 | | Class 2 : distorted nucleosome | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B, ... | | 著者 | Bilokapic, S, Halic, M. | | 登録日 | 2018-02-13 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural rearrangements of the histone octamer translocate DNA.
Nat Commun, 9, 2018
|
|
6FQ8
 
 | |
4FCC
 
 | |
4FHM
 
 | |
4FHL
 
 | | Nucleoporin Nup37 from Schizosaccharomyces pombe | | 分子名称: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Bilokapic, S, Schwartz, T.U. | | 登録日 | 2012-06-06 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I0O
 
 | |
4FHN
 
 | |
2CJB
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with serine | | 分子名称: | CHLORIDE ION, SERINE, SERYL-TRNA SYNTHETASE, ... | | 著者 | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | 登録日 | 2006-03-30 | | 公開日 | 2006-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2CJ9
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with an analog of seryladenylate | | 分子名称: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CHLORIDE ION, SERYL-TRNA SYNTHETASE, ... | | 著者 | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | 登録日 | 2006-03-29 | | 公開日 | 2006-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2CIM
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase | | 分子名称: | CHLORIDE ION, PLATINUM (II) ION, SERYL-TRNA SYNTHETASE, ... | | 著者 | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | 登録日 | 2006-03-24 | | 公開日 | 2006-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2CJA
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | 登録日 | 2006-03-30 | | 公開日 | 2006-06-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
7YAR
 
 | | ZIKV_Fab_G9E | | 分子名称: | E protein, M protein, heavy chain, ... | | 著者 | Shu, B, Thiam-Seng, N, Lok, S. | | 登録日 | 2022-06-28 | | 公開日 | 2023-01-04 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structure and neutralization mechanism of a human antibody targeting a complex Epitope on Zika virus.
Plos Pathog., 19, 2023
|
|
8FU7
 
 | | Structure of Covid Spike variant deltaN135 in fully closed form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | 登録日 | 2023-01-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | 登録日 | 2023-01-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | 登録日 | 2023-01-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6PX1
 
 | | Set2 bound to nucleosome | | 分子名称: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | 著者 | Halic, M, Bilokapic, S. | | 登録日 | 2019-07-24 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6PX3
 
 | | Set2 bound to nucleosome | | 分子名称: | DNA (145-MER), Histone H2B 1.1, Histone H3, ... | | 著者 | Halic, M, Bilokapic, S. | | 登録日 | 2019-07-24 | | 公開日 | 2019-08-28 | | 最終更新日 | 2019-09-04 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6NZO
 
 | | Set2 bound to nucleosome | | 分子名称: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | 著者 | Halic, M, Bilokapic, S. | | 登録日 | 2019-02-14 | | 公開日 | 2019-08-28 | | 最終更新日 | 2019-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
