7PXL
 
 | | X-ray structure of LPMO at 3.6x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ7
 
 | | Structure of an LPMO at 1.13x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYF
 
 | | Structure of LPMO in complex with cellotetraose at 1.39x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ0
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 9.81x10^6 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXK
 
 | | X-ray structure of LPMO at 1.39x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYH
 
 | | Structure of LPMO in complex with cellotetraose at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ3
 
 | | Structure of an LPMO at 5.37x10^3 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXR
 
 | | Room temperature structure of an LPMO. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Meilleur, F, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXJ
 
 | | X-ray structure of LPMO at 5.99x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYG
 
 | | Structure of LPMO in complex with cellotetraose at 3.6x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYM
 
 | | Structure of an LPMO (expressed in E.coli) at 5.61x10^4 Gy | | Descriptor: | Auxiliary activity 9, COPPER (II) ION, SULFATE ION | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYZ
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 2.97x10^6 Gy | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ8
 
 | | Structure of an LPMO at 3.12x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ6
 
 | | Structure of an LPMO at 2.22x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYL
 
 | | Structure of an LPMO (expressed in E.coli) at 1.49x10^4 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ5
 
 | | Structure of an LPMO at 9.56x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6RS7
 
 | | X-ray crystal structure of LsAA9B (deglycosylated form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
6RS9
 
 | | X-ray crystal structure of LsAA9B (xylotetraose soak) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, BICINE, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
6RS8
 
 | | X-ray crystal structure of LsAA9B (transition metals soak) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
6ST6
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant | | Descriptor: | ISOPROPYL ALCOHOL, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Carey, J, Drouard, F, von Wachenfeldt, C, Schulz, A, Linse, S, Lo Leggio, L. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6SZ4
 
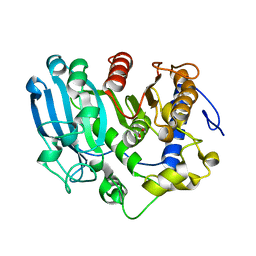 | | The glucuronoyl esterase OtCE15A H408A variant from Opitutus terrae in complex with, and covalently linked to, D-glucuronate | | Descriptor: | FORMIC ACID, beta-D-glucopyranuronic acid, glucuronoyl esterase OtCE15A | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6SYR
 
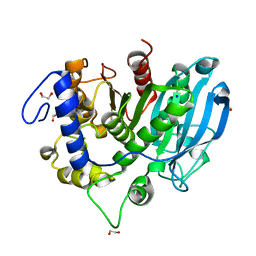 | | The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with D-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6SZ0
 
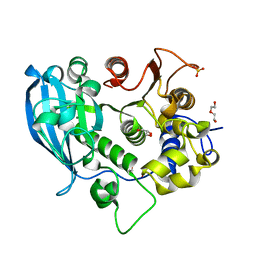 | | The glucuronoyl esterase OtCE15A H408A variant from Opitutus terrae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6T0E
 
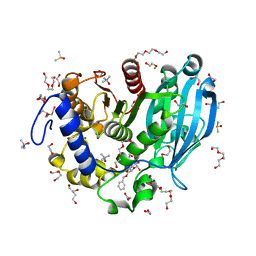 | | The glucuronoyl esterase OtCE15A S267A variant from Opitutus terrae in complex with benzyl D-glucuronoate and D-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-03 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6T0I
 
 | | The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with the aldotetrauronic acid XUX | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-03 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
