6Y93
 
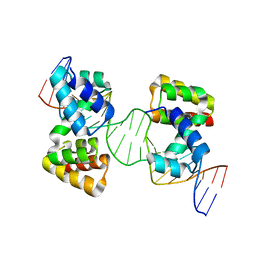 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
1YGS
 
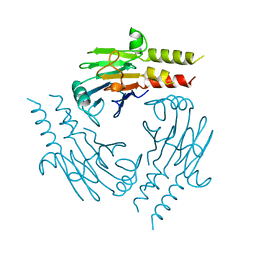 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
2MLH
 
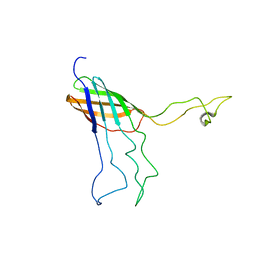 | | NMR Solution Structure of Opa60 from N. Gonorrhoeae in FC-12 Micelles | | Descriptor: | Opacity protein opA60 | | Authors: | Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M, Columbus, L. | | Deposit date: | 2014-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the neisserial outer membrane protein opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2MAF
 
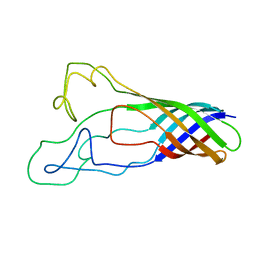 | | Solution structure of opa60 from n. gonorrhoeae | | Descriptor: | Opacity protein opA60 | | Authors: | Columbus, L, Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M. | | Deposit date: | 2013-07-08 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neisserial outer membrane protein Opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
