5HP8
 
 | | Crystal structures of RidA in complex with pyruvate | | Descriptor: | PYRUVIC ACID, Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
5HP7
 
 | | Crystal structures of RidA in the apo form | | Descriptor: | Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
4ANJ
 
 | | MYOSIN VI (MDinsert2-GFP fusion) PRE-POWERSTROKE STATE (MG.ADP.AlF4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Menetrey, J, Isabet, T, Ropars, V, Mukherjea, M, Pylypenko, O, Liu, X, Perez, J, Vachette, P, Sweeney, H.L, Houdusse, A.M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin Vi.
Mol.Cell, 48, 2012
|
|
8WOI
 
 | |
8WOO
 
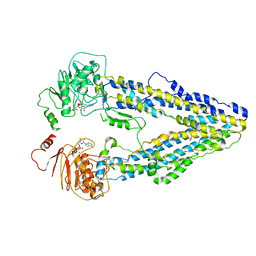 | | Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state | | Descriptor: | ABC transporter B family member 19, Brassinolide, MAGNESIUM ION, ... | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8WP0
 
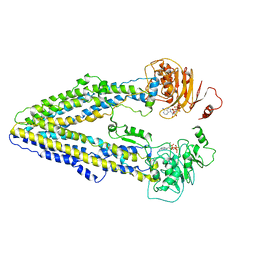 | | Structure of the Arabidopsis E529Q/E1174Q ABCB19 in the ATP bound state | | Descriptor: | ABC transporter B family member 19, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8WOM
 
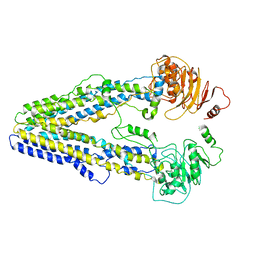 | | Structure of the wild-type Arabidopsis ABCB19 in the brassinolide-bound state | | Descriptor: | ABC transporter B family member 19, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
3HSK
 
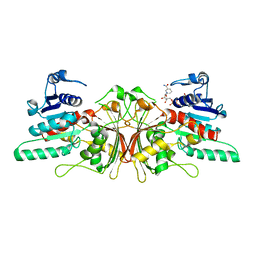 | | Crystal Structure of Aspartate Semialdehyde Dehydrogenase with NADP from Candida albicans | | Descriptor: | Aspartate-semialdehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arachea, B.T, Pavlovsky, A, Liu, X, Viola, R.E. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expansion of the aspartate beta-semialdehyde dehydrogenase family: the first structure of a fungal ortholog.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3J0C
 
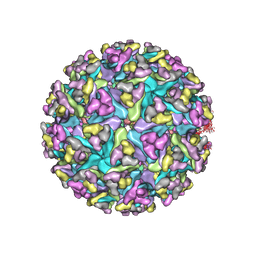 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
3J7L
 
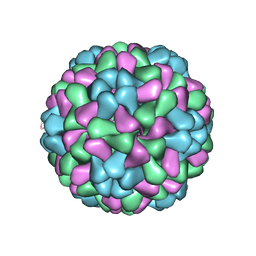 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3L9F
 
 | |
3LBE
 
 | | The Crystal Structure of smu.793 from Streptococcus mutans UA159 bound to acetyl CoA | | Descriptor: | CHLORIDE ION, COENZYME A, Putative uncharacterized protein smu.793 | | Authors: | Su, X.-D, Hou, Q.M, Fan, X.X, Nan, J, Liu, X. | | Deposit date: | 2010-01-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of smu.793 from Streptococcus mutans UA159 bound to acetyl CoA
TO BE PUBLISHED
|
|
3L86
 
 | |
3L9D
 
 | |
3L7Y
 
 | |
3L7V
 
 | |
3LD2
 
 | |
3L7X
 
 | |
3L87
 
 | |
3L9C
 
 | |
3LA8
 
 | |
3LBB
 
 | | The Crystal Structure of smu.793 from Streptococcus mutans UA159 | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein smu.793, SULFATE ION | | Authors: | Su, X.-D, Hou, Q.M, Fan, X.X, Nan, J, Liu, X. | | Deposit date: | 2010-01-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of smu.793 from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3L7W
 
 | |
