2VTF
 
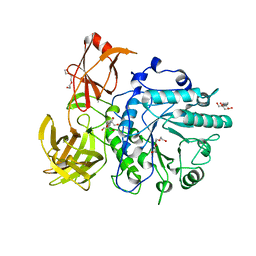 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | Authors: | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
6L3F
 
 | |
6L8K
 
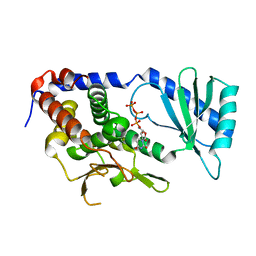 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
8UIQ
 
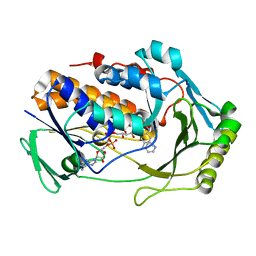 | | H47Q NicC with 2-mercaptopyridine ligand | | Descriptor: | 2-PYRIDINETHIOL, 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hicks, K.A, Perry, K, Turlington, Z.R, Vaz Ferreira de Macedo, S. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RVD
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
