1MTG
 
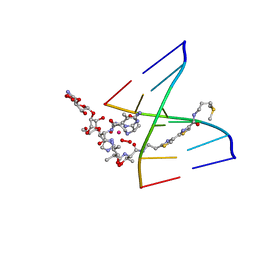 | | NMR Structure of HO2-Co(III)bleomycin A(2) bound to d(GAGCTC)(2) | | Descriptor: | 5'-D(*GP*AP*GP*CP*TP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
3TIW
 
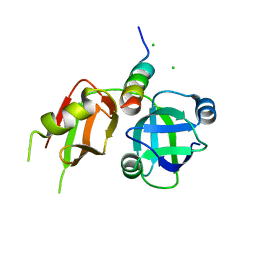 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
1JWA
 
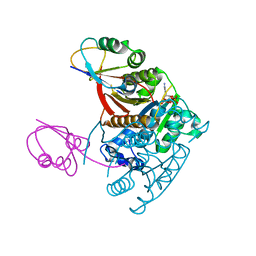 | | Structure of the ATP-bound MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1JWB
 
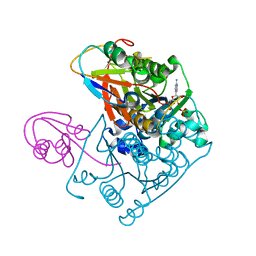 | | Structure of the Covalent Acyl-Adenylate Form of the MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1MXK
 
 | | NMR Structure of HO2-Co(III)bleomycin A(2) Bound to d(GGAAGCTTCC)(2) | | Descriptor: | 5'-D(*GP*GP*AP*AP*GP*CP*TP*TP*CP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-10-02 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3SSP
 
 | |
1ED0
 
 | | NMR structural determination of viscotoxin A3 from Viscum album L. | | Descriptor: | VISCOTOXIN A3 | | Authors: | Romagnoli, S, Ugolini, R, Fogolari, F, Schaller, G, Urech, K, Giannattasio, M, Ragona, L, Molinari, H. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structural determination of viscotoxin A3 from Viscum album L.
Biochem.J., 350, 2000
|
|
3SST
 
 | |
2C0F
 
 | | Structure of Wind Y53F mutant | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3E3I
 
 | |
3SSV
 
 | |
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3SS0
 
 | |
3SSH
 
 | |
3SSL
 
 | |
3SSY
 
 | |
2C0G
 
 | | Structure of PDI-related Chaperone, Wind mutant-Y53S | | Descriptor: | CHLORIDE ION, SODIUM ION, WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3SVC
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: chloride complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
2IBK
 
 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*AP*T)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4PD1
 
 | | Structure of gephyrin E domain with Glycine-beta receptor peptide | | Descriptor: | ACETATE ION, GLYCEROL, Gephyrin, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Modulation of gephyrin-glycine receptor affinity by multivalency.
Acs Chem.Biol., 9, 2014
|
|
3SVE
 
 | |
3SRY
 
 | |
3SVB
 
 | |
3SV5
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
