2BKY
 
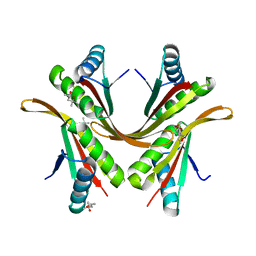 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | 著者 | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | 登録日 | 2005-02-22 | | 公開日 | 2005-07-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
4ES8
 
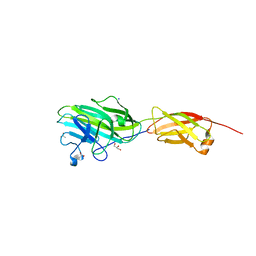 | | Crystal Structure of the adhesin domain of Epf from Streptococcus pyogenes in P212121 | | 分子名称: | ACETATE ION, Epf, GLYCEROL, ... | | 著者 | Linke, C, Siemens, N, Kreikemeyer, B, Baker, E.N. | | 登録日 | 2012-04-23 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | The Extracellular Protein Factor Epf from Streptococcus pyogenes Is a Cell Surface Adhesin That Binds to Cells through an N-terminal Domain Containing a Carbohydrate-binding Module.
J.Biol.Chem., 287, 2012
|
|
4D0A
 
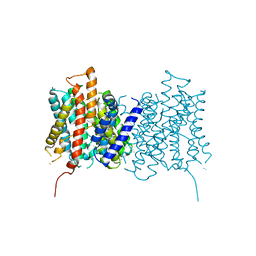 | |
5OYG
 
 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | 分子名称: | Anoctamin-1 | | 著者 | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | 登録日 | 2017-09-08 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
1K4W
 
 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | 分子名称: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | 著者 | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | 登録日 | 2001-10-09 | | 公開日 | 2002-04-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | 分子名称: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | 著者 | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | 登録日 | 2022-05-20 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (4.31 Å) | | 主引用文献 | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | 著者 | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | 登録日 | 2022-05-23 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.38 Å) | | 主引用文献 | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | 分子名称: | Mesothelin, cleaved form, SULFATE ION, ... | | 著者 | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | 登録日 | 2022-05-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
7C2Y
 
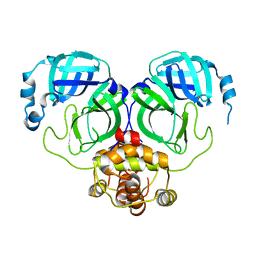 | | The crystal structure of COVID-2019 main protease in the apo state | | 分子名称: | 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-05-10 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | COVID-2019 main protease in the apo state
To Be Published
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | 分子名称: | 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | 登録日 | 2020-05-08 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
6DA1
 
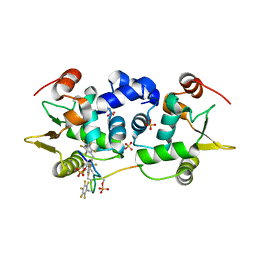 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-01 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.000127 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DAT
 
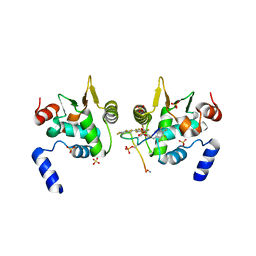 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-02 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35002637 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
8DD7
 
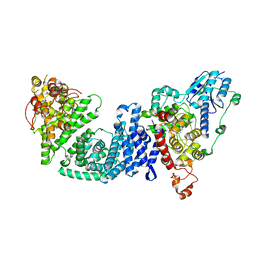 | | The Cryo-EM structure of Drosophila Cryptochrome in complex with Timeless | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Methylated-DNA--protein-cysteine methyltransferase,Cryptochrome-1 fusion, Protein timeless,Methylated-DNA--protein-cysteine methyltransferase fusion | | 著者 | Feng, S, Lin, C, DeOliveira, C.C, Crane, B.R. | | 登録日 | 2022-06-17 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Nature, 617, 2023
|
|
6XIW
 
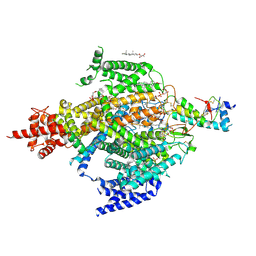 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | 登録日 | 2020-06-22 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-11-25 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
4V9Q
 
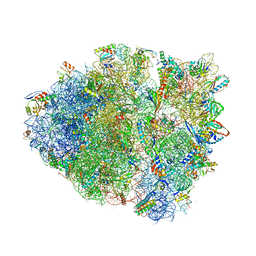 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | 登録日 | 2013-06-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6JJ0
 
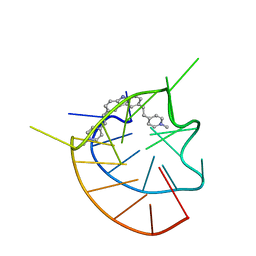 | |
1Y9H
 
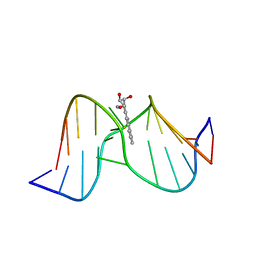 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | 分子名称: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | 著者 | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2004-12-15 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-04-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | 分子名称: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
8J3A
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
