4JF2
 
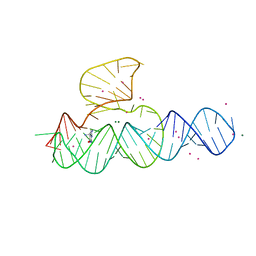 | |
4G6R
 
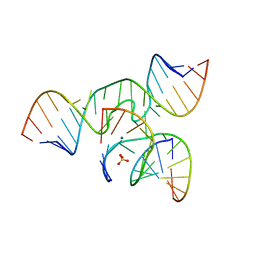 | | Minimal Hairpin Ribozyme in the Transition State with G8I Variation | | 分子名称: | Loop A Ribozyme strand, Loop A Substrate strand, Loop B Ribozyme Strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.832 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6P
 
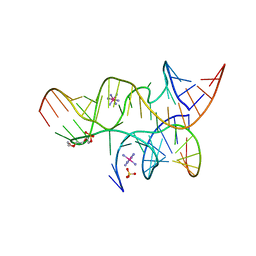 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | 分子名称: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.641 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6S
 
 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | 分子名称: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4RZD
 
 | | Crystal Structure of a PreQ1 Riboswitch | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | 著者 | Wedekind, J.E, Liberman, J.A, Salim, M. | | 登録日 | 2014-12-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D5L
 
 | |
