1F99
 
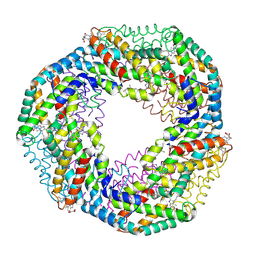 | | CRYSTAL STRUCTURE OF R-PHYCOCYANIN FROM POLYSIPHONIA AT 2.4 A RESOLUTION | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROBILIN, ... | | Authors: | Liang, D.C, Jiang, T, Chang, W.R. | | Deposit date: | 2000-07-09 | | Release date: | 2001-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of R-phycocyanin and possible energy transfer pathways in the phycobilisome.
Biophys.J., 81, 2001
|
|
1LHP
 
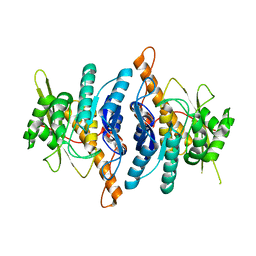 | |
1LHR
 
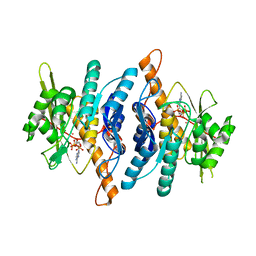 | | Crystal Structure of Pyridoxal Kinase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, POTASSIUM ION, Pyridoxal kinase, ... | | Authors: | Liang, D.C, Jiang, T, Li, M.H. | | Deposit date: | 2002-04-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily
J.BIOL.CHEM., 277, 2002
|
|
1LIA
 
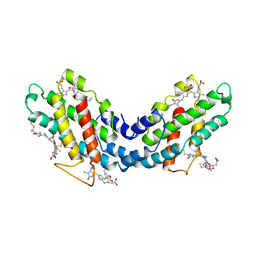 | |
1NP2
 
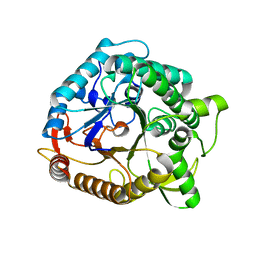 | |
1KN1
 
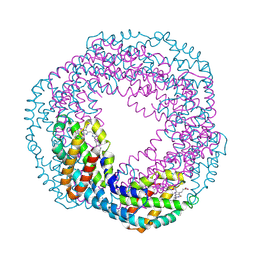 | | Crystal structure of allophycocyanin | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Liang, D.C, Liu, J.Y, Jiang, T, Zhang, J.P, Chang, W.R. | | Deposit date: | 2001-12-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Allophycocyanin from red algae Porphyra yezoensis at 2.2 A resolution
J.BIOL.CHEM., 274, 1999
|
|
1TGR
 
 | | Crystal Structure of mini-IGF-1(2) | | Descriptor: | Insulin-like growth factor IA | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2004-05-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42A crystal structure of mini-IGF-1(2): an analysis of the disulfide isomerization property and receptor binding property of IGF-1 based on the three-dimensional structure
Biochem.Biophys.Res.Commun., 326, 2004
|
|
1RFJ
 
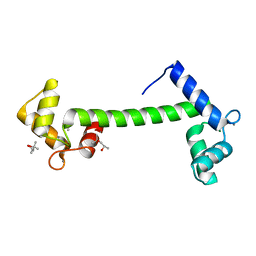 | | Crystal Structure of Potato Calmodulin PCM6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, calmodulin | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of potato calmodulin PCM6: the first report of the three-dimensional structure of a plant calmodulin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4G8B
 
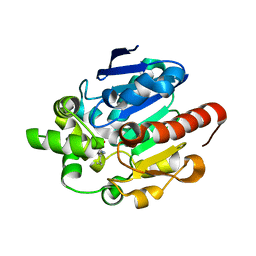 | |
4G9E
 
 | |
4G9G
 
 | |
4G5X
 
 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G8C
 
 | |
4G8D
 
 | |
2PZ0
 
 | | Crystal structure of Glycerophosphodiester Phosphodiesterase (GDPD) from T. tengcongensis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycerophosphoryl diester phosphodiesterase | | Authors: | Shi, L, Liu, J.F, An, X.M, Liang, D.C. | | Deposit date: | 2007-05-17 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism.
Proteins, 72, 2008
|
|
4LTY
 
 | | Crystal Structure of E.coli SbcD at 1.8 A Resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZJ6
 
 | | Crystal structure of human ARL5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Wang, Z.X, Shi, L, Liu, J.F, An, X.M, Chang, W.R, Liang, D.C. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A crystal structure of human ARL5-GDP3'P, a novel member of the small GTP-binding proteins
Biochem.Biophys.Res.Commun., 332, 2005
|
|
1JR9
 
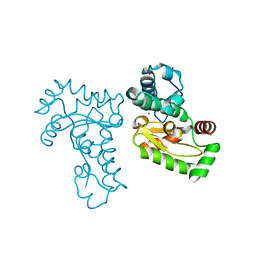 | | Crystal Structure of manganese superoxide dismutases from Bacillus halodenitrificans | | Descriptor: | MANGANESE (II) ION, ZINC ION, manganese superoxide dismutase | | Authors: | Liao, J, Liu, M.Y, Chang, T, Li, M, LeGall, J, Gui, L.L, Zhang, J.P, Jiang, T, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-08-13 | | Release date: | 2002-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of manganese superoxide dismutase from Bacillus halodenitrificans, a component of the so-called "green protein".
J.Struct.Biol., 139, 2002
|
|
1KCB
 
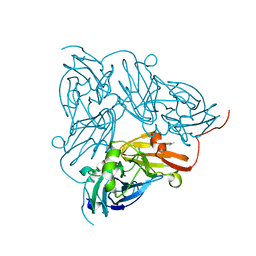 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
3VE5
 
 | |
3VDU
 
 | |
3VDP
 
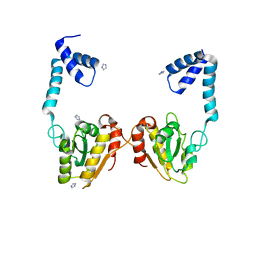 | |
1SQW
 
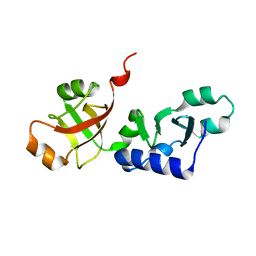 | | Crystal structure of KD93, a novel protein expressed in the human pro | | Descriptor: | Saccharomyces cerevisiae Nip7p homolog | | Authors: | Liu, J.F, Wang, X.Q, Wang, Z.X, Chen, J.R, Jiang, T, An, X.M, Chan, W.R, Liang, D.C. | | Deposit date: | 2004-03-19 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of KD93, a novel protein expressed in human hematopoietic stem/progenitor cells.
J.Struct.Biol., 148, 2004
|
|
2QDL
 
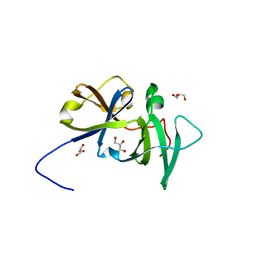 | |
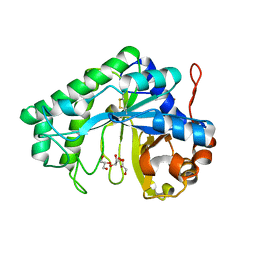
2QHA
 
 | |
