1VMP
 
 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
5JYT
 
 | |
7N82
 
 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
6UF2
 
 | |
2J48
 
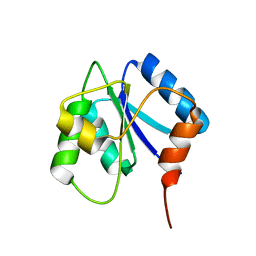 | |
1DK9
 
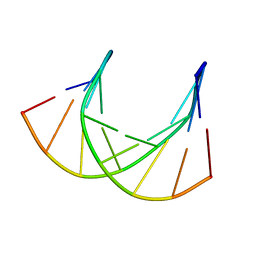 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1DK6
 
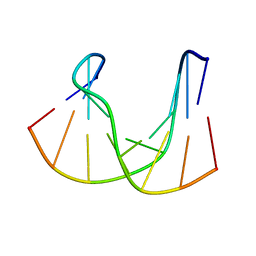 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
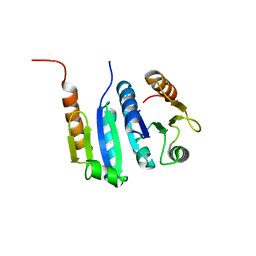
5JYV
 
 | |
5JYU
 
 | |
1M2F
 
 | |
1R8J
 
 | |
1M2E
 
 | |
1Q6A
 
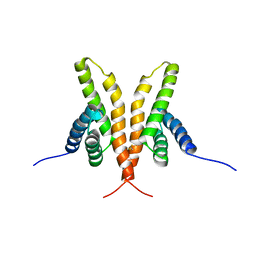 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q6B
 
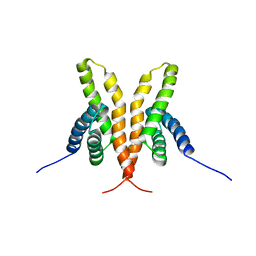 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Ensemble of 25 Structures | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Golden, S.S, Holzenburg, A, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5JWQ
 
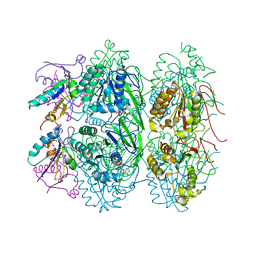 | | Crystal structure of KaiC S431E in complex with foldswitch-stabilized KaiB from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.871 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5JWO
 
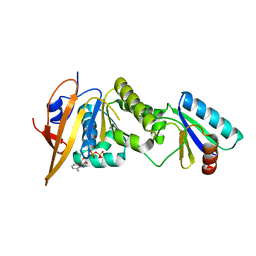 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
1T4Z
 
 | |
1SV1
 
 | |
1T4Y
 
 | |
5JWR
 
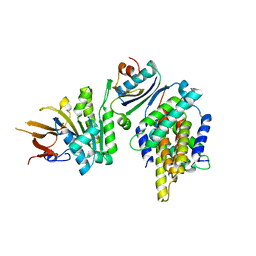 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC and a dimer of KaiA C-terminal domains from Thermosynechococcus elongatus | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC, ... | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y.G, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
1SUY
 
 | |
