5HSO
 
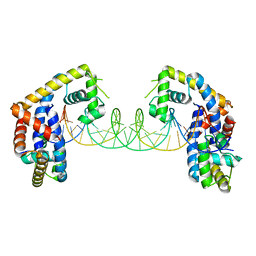 | | Crystal structure of MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN Rv2887 complex with DNA | | Descriptor: | DNA (30-MER), the upstream sequence of Rv0560c, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5HQP
 
 | | Crystal structure of the ERp44-peroxiredoxin 4 complex | | Descriptor: | Endoplasmic reticulum resident protein 44, Peroxiredoxin-4 | | Authors: | Yang, K, Li, D.F, Wang, X, Wang, C.C. | | Deposit date: | 2016-01-22 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the ERp44-Peroxiredoxin 4 Complex Reveals the Molecular Mechanisms of Thiol-Mediated Protein Retention.
Structure, 24, 2016
|
|
3VYN
 
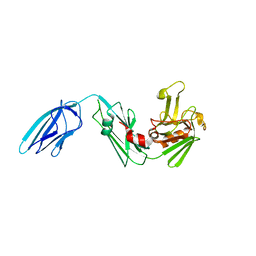 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2 N55 truncation mutant (resideus 55-408) | | Descriptor: | Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
3VYP
 
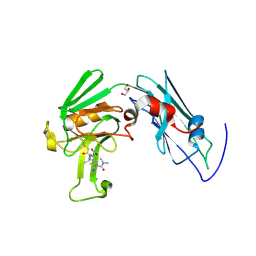 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2-N140 adduct with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
3VYO
 
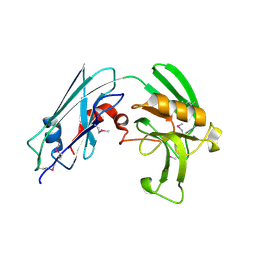 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2 N140 truncation mutant (resideus 140-408) | | Descriptor: | Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
5X7Z
 
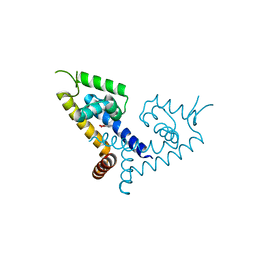 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 COMPLEX WITH P-AMINOSALICYLIC ACID | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5Y6G
 
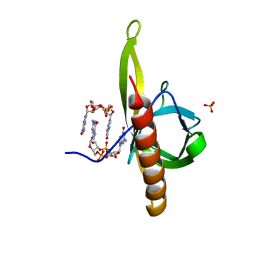 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
5Y6H
 
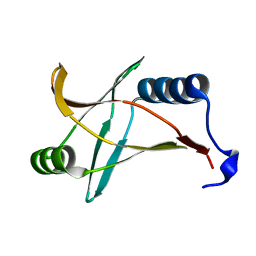 | |
5Y6F
 
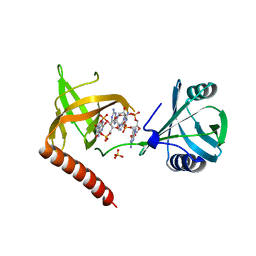 | | Crystal structure of YcgR in complex with c-di-GMP from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
5YT1
 
 | |
5YZ2
 
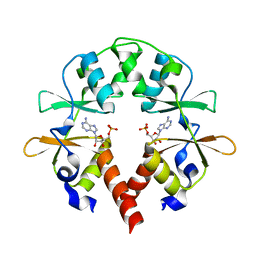 | | the cystathionine-beta-synthase (CBS) domain of magnesium and cobalt efflux protein CorC in complex with both C2'- and C3'-endo AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Feng, N, Qi, C, Li, D.F, Wang, D.C. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C2'- and C3'-endo equilibrium for AMP molecules bound in the cystathionine-beta-synthase domain.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5Z2L
 
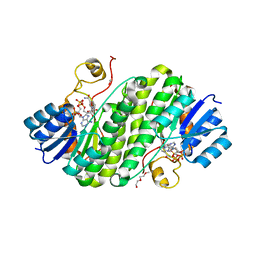 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
8WDF
 
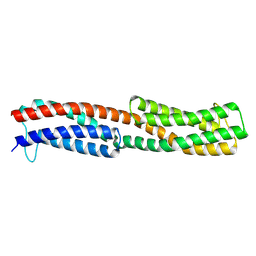 | | Chemoreceptor PilJ from Pseudomonas aeruginosa PA14 | | Descriptor: | Chemotaxis chemoreceptor PilJ | | Authors: | Cui, R, Li, D.F. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | The ligand binding domain of a type IV pilus chemoreceptor PilJ has a different fold from that of another PilJ-type receptor McpN.
Biochem.Biophys.Res.Commun., 706, 2024
|
|
6IYV
 
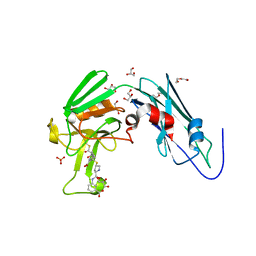 | | Crystal sturucture of L,D-transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with ertapenem adduct | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhao, F, Li, D.F, Wang, D.C. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1-beta-methyl group confers a lower affinity of l,d-transpeptidase LdtMt2for ertapenem than for imipenem.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
3AYC
 
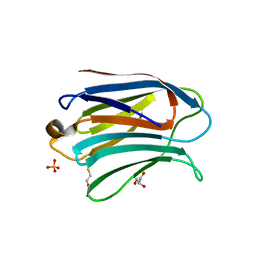 | | Crystal structure of galectin-3 CRD domian complexed with GM1 pentasaccharide | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Galectin-3, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3AYE
 
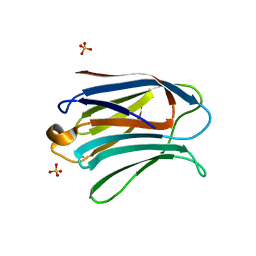 | |
3AYA
 
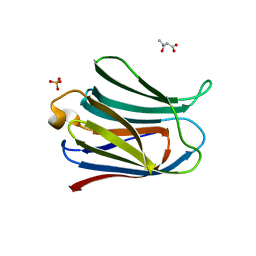 | | Crystal structure of galectin-3 CRD domian complexed with Thomsen-Friedenreich antigen | | Descriptor: | Galectin-3, SULFATE ION, THREONINE, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3AYD
 
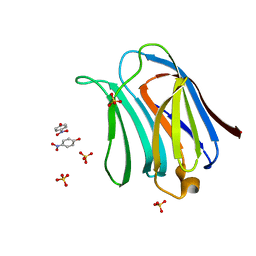 | | Crystal structure of galectin-3 CRD domian complexed with TFN | | Descriptor: | Galectin-3, P-NITROPHENOL, SULFATE ION, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3TC1
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
3W53
 
 | |
3WMS
 
 | | The crystal structure of Y195I mutant alpha-cyclodextrin glycosyltransferase from Paenibacillus macerans | | Descriptor: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | Authors: | Xie, T, Hou, Y.J, Li, D.F, Yue, Y, Qian, S.J, Chao, Y.P. | | Deposit date: | 2013-11-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of a mutant Y195I alpha-cyclodextrin glycosyltransferase with switched product specificity from alpha-cyclodextrin to beta-/ gamma-cyclodextrin
J.Biotechnol., 182-183, 2014
|
|
3X3M
 
 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P212121 | | Descriptor: | CALCIUM ION, ESX-1 secretion system protein EccB1 | | Authors: | Zhang, X.L, Li, D.F, Zhang, X.E, Bi, L.J, Wang, D.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Core component EccB1 of the Mycobacterium tuberculosis type VII secretion system is a periplasmic ATPase.
Faseb J., 29, 2015
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
