1LME
 
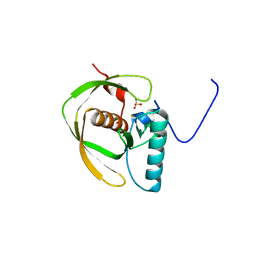 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM6
 
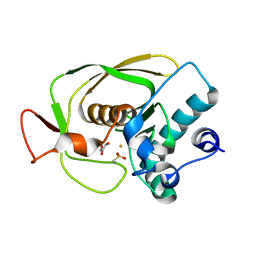 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1N5N
 
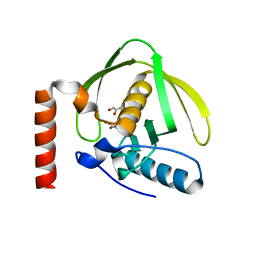 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1SJ5
 
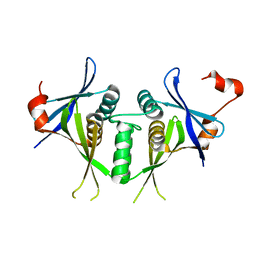 | | Crystal structure of a duf151 family protein (tm0160) from thermotoga maritima at 2.8 A resolution | | Descriptor: | conserved hypothetical protein TM0160 | | Authors: | Spraggon, G, Panatazatos, D, Klock, H.E, Wilson, I.A, Woods Jr, V.L, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-03-02 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM1171.
Protein Sci., 13, 2004
|
|
1S96
 
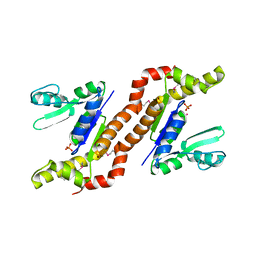 | | The 2.0 A X-ray structure of Guanylate Kinase from E.coli | | Descriptor: | Guanylate kinase, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Kreusch, A, Spraggon, G, Klock, H.E, McMullan, D, Vincent, J, Rodrigues, K, Lesley, S.A. | | Deposit date: | 2004-02-03 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Guanylate Kinase from Escherichia coli at 2.0 A resolution
To be Published
|
|
1R73
 
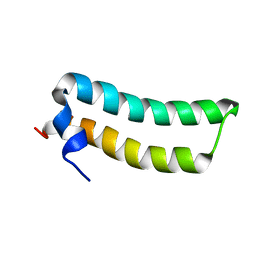 | | Solution Structure of TM1492, the L29 ribosomal protein from Thermotoga maritima | | Descriptor: | 50S ribosomal protein L29 | | Authors: | Peti, W, Etezady-Esfarjani, T, Herrmann, T, Klock, H.E, Lesley, S.A, Wuethrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-10-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR for structural proteomics of Thermotoga maritima: Screening and structure determination
J.STRUCT.FUNCT.GENOM., 5, 2004
|
|
1RDU
 
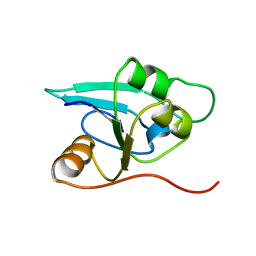 | | NMR STRUCTURE OF A PUTATIVE NIFB PROTEIN FROM THERMOTOGA (TM1290), WHICH BELONGS TO THE DUF35 FAMILY | | Descriptor: | conserved hypothetical protein | | Authors: | Etezady-Esfarjani, T, Herrmann, T, Peti, W, Klock, H.E, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determination of the Hypothetical Protein TM1290 from Thermotoga Maritima using Automated NOESY Analysis.
J.Biomol.NMR, 29, 2004
|
|
3RTA
 
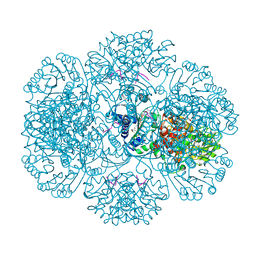 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Acetyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RT9
 
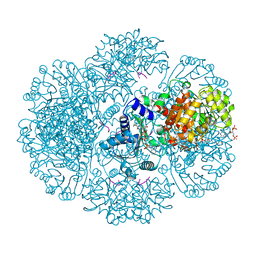 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A | | Descriptor: | COENZYME A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTG
 
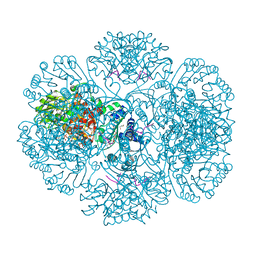 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSF
 
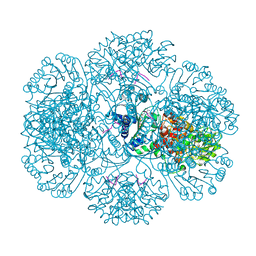 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTB
 
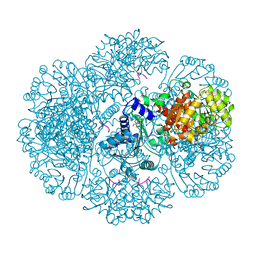 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Adenosine-3'-5'-Diphosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRE
 
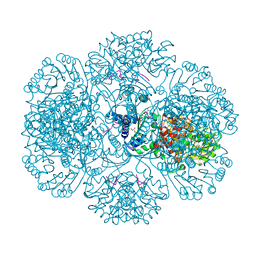 | | Crystal Structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSS
 
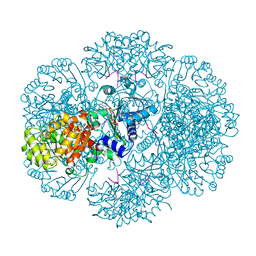 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTC
 
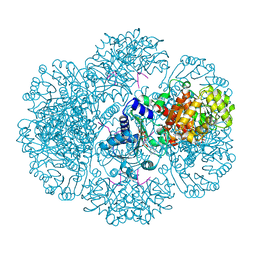 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRF
 
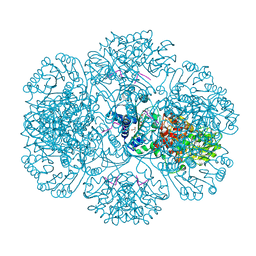 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTD
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH and ADP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRB
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRJ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RT7
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS8
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTE
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSQ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
