1SRK
 
 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
1ZM1
 
 | | Crystal structures of complex F. succinogenes 1,3-1,4-beta-D-glucanase and beta-1,3-1,4-cellotriose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tsai, L.C, Shyur, L.F, Cheng, Y.S, Lee, S.H. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase in complex with beta-1,3-1,4-cellotriose
J.Mol.Biol., 354, 2005
|
|
2EYI
 
 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.7 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
2EYN
 
 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.8 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
4NTZ
 
 | | Crystal structure of Adenylate kinase from Streptococcus pneumoniae | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Thach, T.T, Luong, T.T, Lee, S.H, Rhee, D.K. | | Deposit date: | 2013-12-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Adenylate kinase from Streptococcus pneumoniae is essential for growth through its catalytic activity
FEBS Open Bio, 4, 2014
|
|
4NU0
 
 | | Crystal structure of Adenylate kinase from Streptococcus pneumoniae with Ap5A | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Thach, T.T, Luong, T.T, Lee, S.H, Rhee, D.K. | | Deposit date: | 2013-12-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.485 Å) | | Cite: | Adenylate kinase from Streptococcus pneumoniae is essential for growth through its catalytic activity
FEBS Open Bio, 4, 2014
|
|
5SV6
 
 | |
5XM3
 
 | | Crystal Structure of Methanol dehydrogenase from Methylophaga aminisulfidivorans | | Descriptor: | Glucose dehydrogenase, MAGNESIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Cao, T.P, Choi, J.M, Lee, S.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT
J. Microbiol., 56, 2018
|
|
5GLD
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLA
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR10 containing 10 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GL9
 
 | | Crystal structure of the class A beta-lactamase PenL | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLB
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLC
 
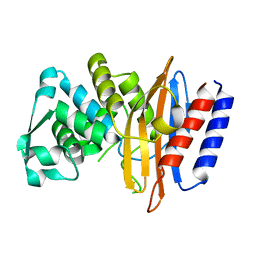 | | Crystal structure of the class A beta-lactamase PenL-tTR11 containing 20 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
7C90
 
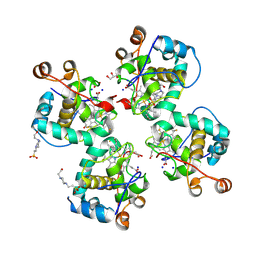 | | Crystal structure of Cytochrome CL from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT (Ma-CytcL) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Cytochrome c, ... | | Authors: | Ghosh, S, Dhanasingh, I, Lee, S.H. | | Deposit date: | 2020-06-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of CytochromecLfrom the Aquatic Methylotrophic BacteriumMethylophaga aminisulfidivoransMPT.
J Microbiol Biotechnol., 30, 2020
|
|
7CVX
 
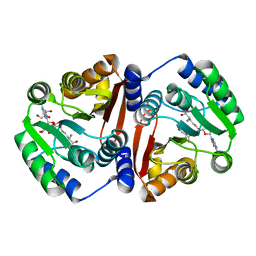 | |
7CMY
 
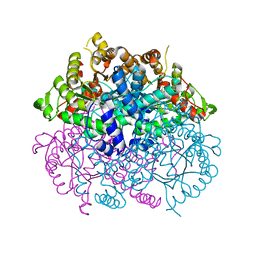 | | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate | | Descriptor: | GLYOXYLIC ACID, Isocitrate lyase, MAGNESIUM ION, ... | | Authors: | Kim, K, Ki, D, Lee, S.H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate
To Be Published
|
|
7CMX
 
 | |
7CVW
 
 | |
7CVV
 
 | |
7CVU
 
 | |
