5JV4
 
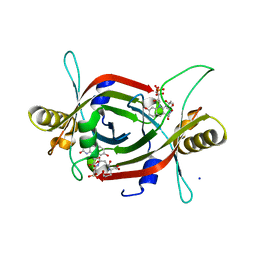 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis with F420 bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COENZYME F420, ... | | Authors: | Lee, B.M, Carr, P.D, Jackson, C.J. | | Deposit date: | 2016-05-10 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis with F420 bound
To Be Published
|
|
4ZKY
 
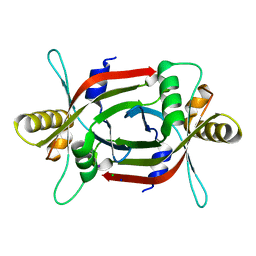 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, IODIDE ION, Pyridoxamine 5-phosphate oxidase, ... | | Authors: | Lee, B.M, Carr, P.D, Ahmed, F.H, Jackson, C.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
2HGH
 
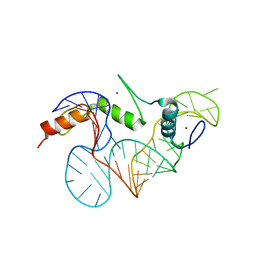 | |
7KL8
 
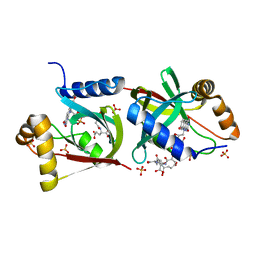 | | Structure of F420 binding protein Rv1558 from Mycobacterium tuberculosis with F420 bound | | Descriptor: | COENZYME F420, COENZYME F420-3, Deazaflavin-dependent nitroreductase, ... | | Authors: | Lee, B.M, Tan, L.L, Jackson, C.J. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Potency boost of a Mycobacterium tuberculosis dihydrofolate reductase inhibitor by multienzyme F 420 H 2 -dependent reduction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2JOX
 
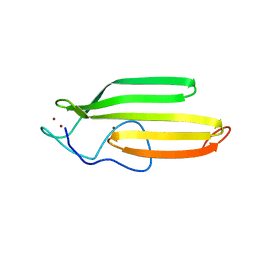 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2M13
 
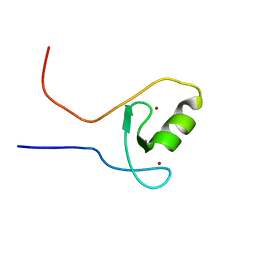 | | The ZZ domain of cytoplasmic polyadenylation element binding protein 1 (CPEB1) | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 1, ZINC ION | | Authors: | Lee, B.M, Merkel, D.J, Wells, S.B, Hilburn, B.C, Elazzouzi, F, Perez-Alvarado, G.C. | | Deposit date: | 2012-11-14 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Region of Cytoplasmic Polyadenylation Element Binding Protein Is a ZZ Domain with Potential for Protein-Protein Interactions.
J.Mol.Biol., 425, 2013
|
|
3E5N
 
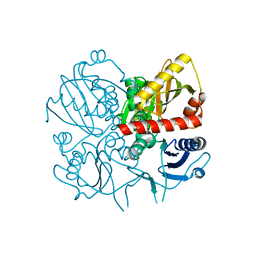 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
3E6G
 
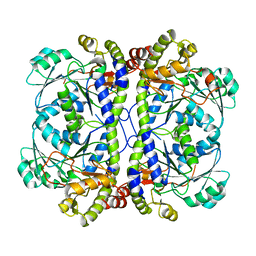 | | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae | | Descriptor: | Cystathionine gamma-lyase-like protein | | Authors: | Ngo, H.P.T, Kim, J.K, Kim, H.S, Jung, J.H, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, H.W, Kang, L.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
To be published
|
|
2PRT
 
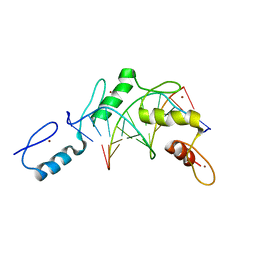 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*CP*GP*CP*CP*CP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*GP*GP*GP*CP*GP*TP*CP*TP*G)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
1MFS
 
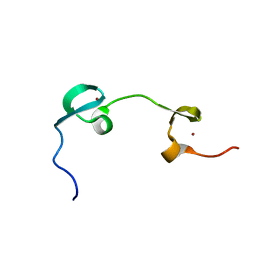 | | DYNAMICAL BEHAVIOR OF THE HIV-1 NUCLEOCAPSID PROTEIN; NMR, 30 STRUCTURES | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Turner, B.G, De Guzman, R.N, Lee, B.M, Tjandra, N. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dynamical behavior of the HIV-1 nucleocapsid protein.
J.Mol.Biol., 279, 1998
|
|
1AFV
 
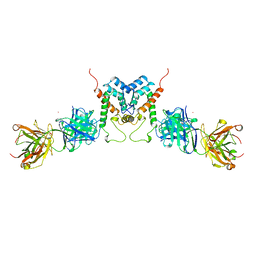 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
1GWP
 
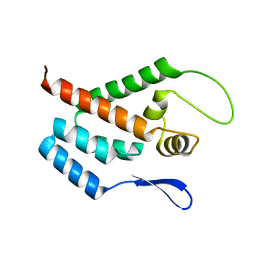 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
2JPA
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
4ME6
 
 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
