1DUD
 
 | |
1BW5
 
 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | Descriptor: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | Authors: | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
2LVF
 
 | | Solution structure of the Brazil Nut 2S albumin Ber e 1 | | Descriptor: | 2S albumin | | Authors: | Rundqvist, L, Tengel, T, Zdunek, J, Schleucher, J, Alcocer, M.J, Larsson, G. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, copper binding and backbone dynamics of recombinant Ber e 1-the major allergen from Brazil nut.
Plos One, 7, 2012
|
|
1OP9
 
 | | Complex of human lysozyme with camelid VHH HL6 antibody fragment | | Descriptor: | HL6 camel VHH fragment, Lysozyme C | | Authors: | Dumoulin, M, Last, A.M, Desmyter, A, Decanniere, K, Canet, D, Larsson, G, Spencer, A, Archer, D.B, Sasse, J, Muyldermans, S, Wyns, L, Redfield, C, Matagne, A, Robinson, C.V, Dobson, C.M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme
Nature, 424, 2003
|
|
1M56
 
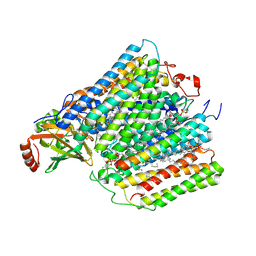 | | Structure of cytochrome c oxidase from Rhodobactor sphaeroides (Wild Type) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
1M57
 
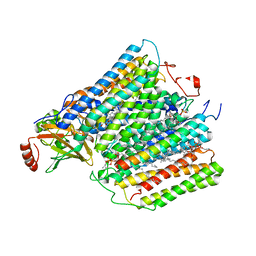 | | Structure of cytochrome c oxidase from Rhodobacter sphaeroides (EQ(I-286) mutant)) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
1EUW
 
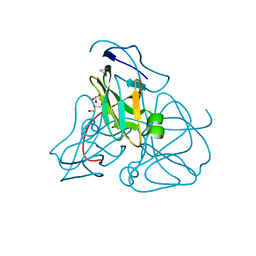 | | ATOMIC RESOLUTION STRUCTURE OF E. COLI DUTPASE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, ETHYL MERCURY ION, GLYCEROL | | Authors: | Gonzalez, A, Cedergren, E, Larsson, G, Persson, R. | | Deposit date: | 2000-04-17 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EU5
 
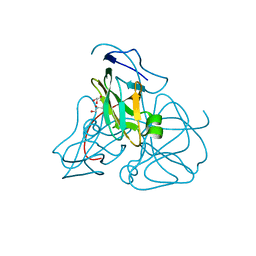 | | STRUCTURE OF E. COLI DUTPASE AT 1.45 A | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, GLYCEROL | | Authors: | Gonzalez, A, Larsson, G, Persson, R, Cedergren-Zeppezauer, E. | | Deposit date: | 2000-04-13 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DUP
 
 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
1VJM
 
 | | Deformation of helix C in the low-temperature L-intermediate of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Edman, K, Royant, A, Larsson, G, Jacobson, F, Taylor, T, van der Spoel, D, Landau, E.M, Pebay-Peyroula, E, Neutze, R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin.
J.Biol.Chem., 279, 2004
|
|
