1LRR
 
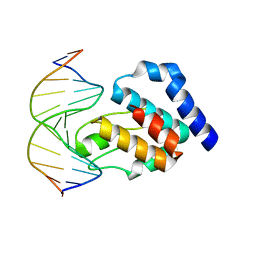 | | CRYSTAL STRUCTURE OF E. COLI SEQA COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*GP*TP*CP*GP*(6MA)P*TP*CP*GP*GP*TP*G)-3', 5'-D(*CP*AP*CP*CP*GP*AP*TP*CP*GP*AP*CP*T)-3', SeqA protein | | Authors: | Guarne, A, Zhao, Q, Guirlando, R, Yang, W. | | Deposit date: | 2002-05-15 | | Release date: | 2002-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex
NAT.STRUCT.BIOL., 9, 2002
|
|
1MN3
 
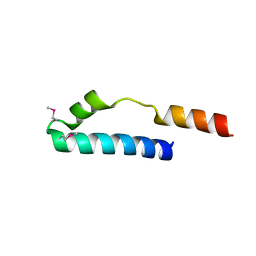 | | Cue domain of yeast Vps9p | | Descriptor: | Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2002-09-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p
Cell(Cambridge,Mass.), 113, 2003
|
|
6TLZ
 
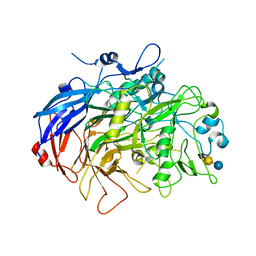 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 3'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
7SO8
 
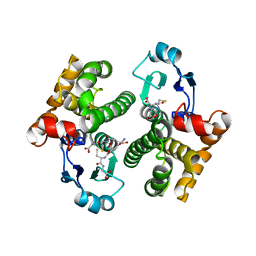 | |
6TM0
 
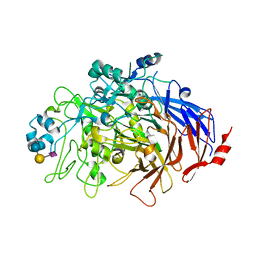 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
6RCC
 
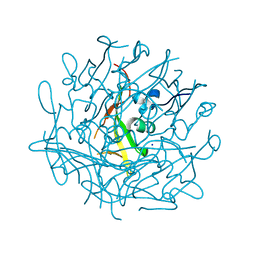 | | Domain C P140 Mycoplasma genitalium | | Descriptor: | Adhesin P1, CHLORIDE ION, SODIUM ION | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6RCD
 
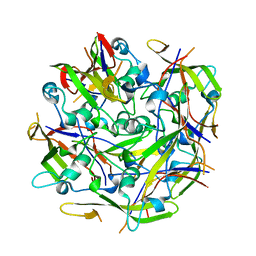 | | Octamer C-Domain P140 Mycoplasma genitalium. | | Descriptor: | MgPa adhesin | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7UXE
 
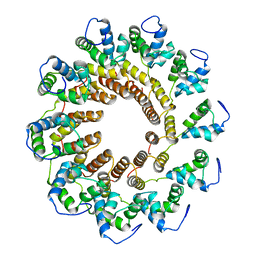 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
2J4E
 
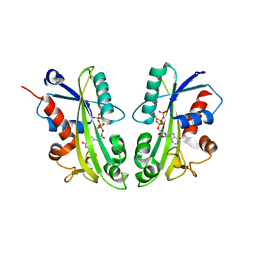 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
6IAR
 
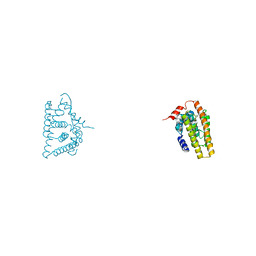 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | Descriptor: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | Authors: | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
2GF9
 
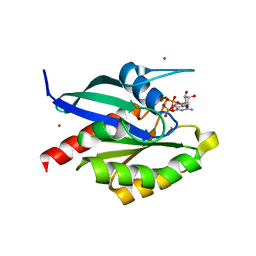 | | Crystal structure of human RAB3D in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-3D, ... | | Authors: | Hong, B, Wang, J, Shen, L, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of human RAB3D in complex with GDP
To be Published
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2GRY
 
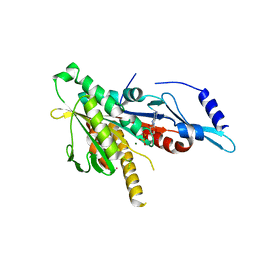 | | Crystal structure of the human KIF2 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human KIF2 motor domain in complex with ADP
to be published
|
|
2E2D
 
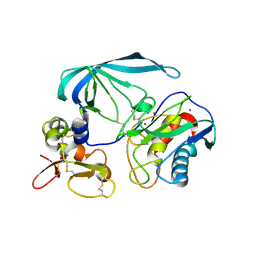 | | Flexibility and variability of TIMP binding: X-ray structure of the complex between collagenase-3/MMP-13 and TIMP-2 | | Descriptor: | CALCIUM ION, Matrix metallopeptidase 13, Metalloproteinase inhibitor 2, ... | | Authors: | Maskos, K, Lang, R, Tschesche, H, Bode, W. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility and Variability of TIMP Binding: X-ray Structure of the Complex Between Collagenase-3/MMP-13 and TIMP-2
J.Mol.Biol., 366, 2007
|
|
3J9Z
 
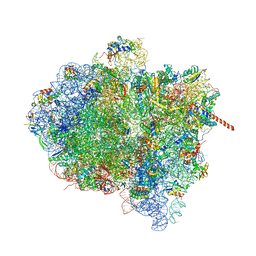 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
2OCB
 
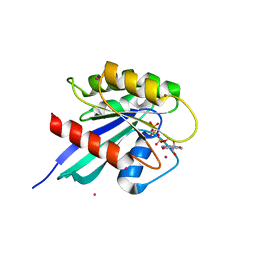 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
3DLT
 
 | |
6B3U
 
 | | Solution Structure of HIV-1 GP41 Transmembrane Domain in Bicelles | | Descriptor: | HIV-1 GP41 Transmembrane Domain | | Authors: | Chiliveri, S.C, Louis, J.M, Ghirlando, R, Baber, J.L, Bax, A. | | Deposit date: | 2017-09-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tilted, Uninterrupted, Monomeric HIV-1 gp41 Transmembrane Helix from Residual Dipolar Couplings.
J. Am. Chem. Soc., 140, 2018
|
|
3JA1
 
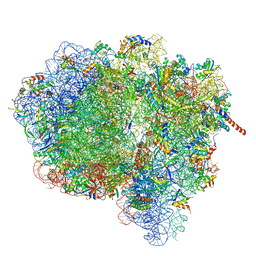 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
6BJS
 
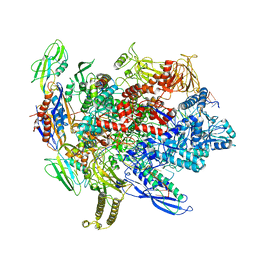 | | CryoEM structure of E.coli his pause elongation complex without pause hairpin | | Descriptor: | DNA (32-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Landick, R, Darst, S.A. | | Deposit date: | 2017-11-06 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.
Mol. Cell, 69, 2018
|
|
3DKR
 
 | |
6C6S
 
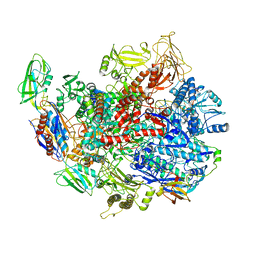 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6U
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6QSQ
 
 | |
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | Descriptor: | Huntingtin | | Authors: | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
