2PK6
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10033 | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl- L-cysteinyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
2PK5
 
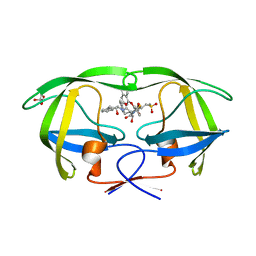 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I ) in Complex with KNI-10075 | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-3-(methyl sulfonyl)-L-alanyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
