7QP2
 
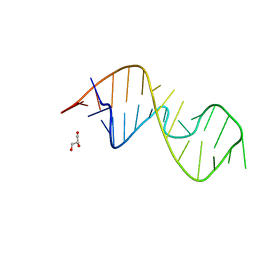 | | 1-deazaguanosine modified-RNA Sarcin Ricin Loop | | 分子名称: | GLYCEROL, RNA (27-MER) | | 著者 | Ennifar, E, Micura, R, Bereiter, R, Renard, E, Kreutz, C. | | 登録日 | 2021-12-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | 1-Deazaguanosine-Modified RNA: The Missing Piece for Functional RNA Atomic Mutagenesis.
J.Am.Chem.Soc., 144, 2022
|
|
6GZK
 
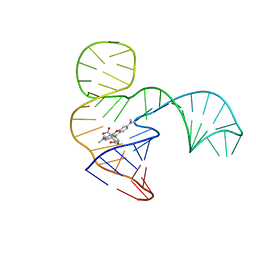 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | 分子名称: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | 著者 | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | 登録日 | 2018-07-04 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
8PXS
 
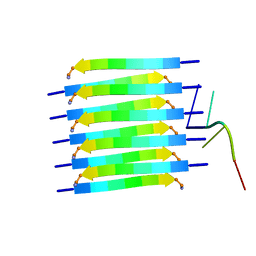 | | Short RNA binding to peptide amyloids | | 分子名称: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | 著者 | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
6GZR
 
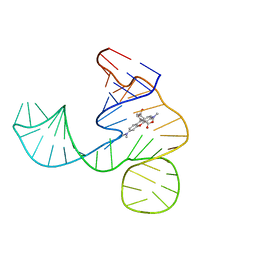 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | 分子名称: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | 著者 | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | 登録日 | 2018-07-05 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
3CJZ
 
 | | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m22G:A pairs | | 分子名称: | RNA (5'-R(*GP*GP*AP*CP*GP*(M2G)P*AP*CP*GP*UP*CP*CP*U)-3') | | 著者 | Pallan, P.S, Kreutz, C, Bosio, S, Micura, R, Egli, M. | | 登録日 | 2008-03-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G:A pairs.
Rna, 14, 2008
|
|
5NPA
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | 分子名称: | Loquacious | | 著者 | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | 登録日 | 2017-04-16 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | 分子名称: | Loquacious, isoform F | | 著者 | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | 登録日 | 2017-04-16 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | 分子名称: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | 著者 | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | 登録日 | 2016-09-17 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
