7VC7
 
 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
7VC6
 
 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, xylan 1,4-beta-xylosidase | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR7
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
2J2M
 
 | | Crystal Structure Analysis of Catalase from Exiguobacterium oxidotolerans | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hara, I, Ichise, N, Kojima, K, Kondo, H, Ohgiya, S, Matsuyama, H, Yumoto, I. | | Deposit date: | 2006-08-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Relationship between the Size of the Bottleneck 15 a from Iron in the Main Channel and the Reactivity of Catalase Corresponding to the Molecular Size of Substrates.
Biochemistry, 46, 2007
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Improvement of substrate specificity of the direct electron transfer type FAD-dependent glucose dehydrogenase catalytic subunit.
J.Biotechnol., 2024
|
|
4YNT
 
 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
4YNU
 
 | | Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone | | Descriptor: | D-glucono-1,5-lactone, FLAVIN-ADENINE DINUCLEOTIDE, Glucose oxidase, ... | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
6A2U
 
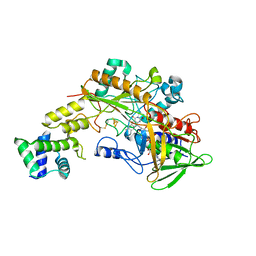 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Kojima, K, Yoshimatsu, K, Shiota, M, Yamazaki, T, Ferri, S, Tsugawa, W, Kamitori, S, Sode, K. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1HR6
 
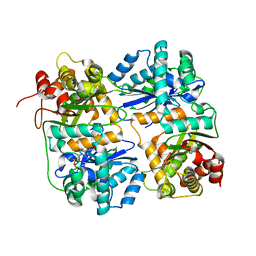 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
6KJL
 
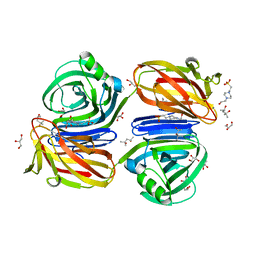 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6KFQ
 
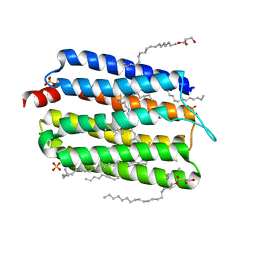 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
