4NFW
 
 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | Descriptor: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | Authors: | Hong, M.K, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-14 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UU0
 
 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UXI
 
 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3E5N
 
 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
3E6G
 
 | | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae | | Descriptor: | Cystathionine gamma-lyase-like protein | | Authors: | Ngo, H.P.T, Kim, J.K, Kim, H.S, Jung, J.H, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, H.W, Kang, L.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae
To be published
|
|
6NJR
 
 | | Spin-Labeled T177C/A637C Mutant of Rat CYPOR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.K. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Studies of the Membrane-Binding Domain of NADPH-Cytochrome P450 Oxidoreductase.
Biochemistry, 58, 2019
|
|
3RFC
 
 | |
3R5F
 
 | |
6KM4
 
 | | Human Carbonic Anhydrase II native 07 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM1
 
 | | Human Carbonic Anhydrase II V143I variant 13 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KLZ
 
 | | Human Carbonic Anhydrase II V143I variant 00 atm CO2 | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM5
 
 | | Human Carbonic Anhydrase II native 13 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM3
 
 | | Human Carbonic Anhydrase II native 00 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM0
 
 | | Human Carbonic Anhydrase II V143I variant 07 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM2
 
 | | Human Carbonic Anhydrase II V143I variant 15 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM6
 
 | | Human Carbonic Anhydrase II native 15 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
4NT8
 
 | | Formyl-methionine-alanine complex structure of peptide deformylase from Xanthomoonas oryzae pv. oryzae | | Descriptor: | ACETATE ION, ALANINE, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-12-02 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure of Xoo1075, a peptide deformylase, from Xanthomonas oryzae pv. oryzae
To be Published
|
|
4ME6
 
 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
7L18
 
 | | Crystal structure of a tandem deletion mutant of rat NADPH-cytochrome P450 reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Xia, C, Shen, A.L, Kim, J.J.K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Structural and kinetic investigations of the carboxy terminus of NADPH-cytochrome P450 oxidoreductase.
Arch.Biochem.Biophys., 701, 2021
|
|
3JTQ
 
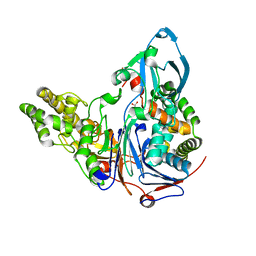 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JTR
 
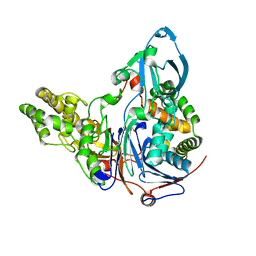 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
7Y2M
 
 | |
7Y2C
 
 | |
7Y2E
 
 | |
