1IYM
 
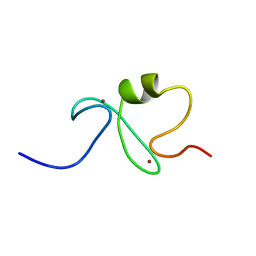 | | RING-H2 finger domain of EL5 | | Descriptor: | EL5, ZINC ION | | Authors: | Katoh, E, Katoh, S, Minami, E, Yamazaki, T. | | Deposit date: | 2002-08-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
J.Biol.Chem., 278, 2003
|
|
1DCJ
 
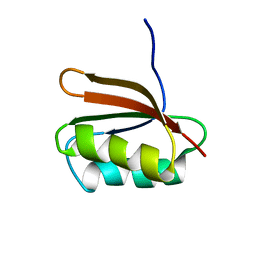 | | SOLUTION STRUCTURE OF YHHP, A NOVEL ESCHERICHIA COLI PROTEIN IMPLICATED IN THE CELL DIVISION | | Descriptor: | YHHP PROTEIN | | Authors: | Katoh, E, Hatta, T, Shindo, H, Mizuno, T, Yamazaki, T. | | Deposit date: | 1999-11-05 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division.
J.Mol.Biol., 304, 2000
|
|
3WRW
 
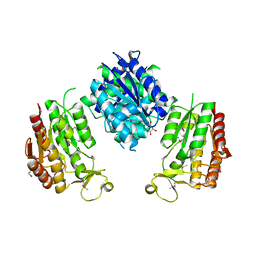 | |
2ZOM
 
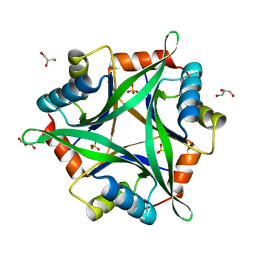 | | Crystal structure of CutA1 from Oryza sativa | | Descriptor: | GLYCEROL, Protein CutA, chloroplast, ... | | Authors: | Kezuka, Y, Bagautdinov, B, Katoh, S, Ohtake, Y, Yutani, K, Nonaka, T, Katoh, E. | | Deposit date: | 2008-05-23 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of CutA1 from Oryza sativa
To be Published
|
|
1TH5
 
 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU1 | | Authors: | Kumeta, H, Ogura, K, Asayama, M, Katoh, S, Katoh, E, Inagaki, F. | | Deposit date: | 2004-06-01 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the domain II of a chloroplastic NifU-like protein OsNifU1A.
J.Biomol.Nmr, 38, 2007
|
|
2JNV
 
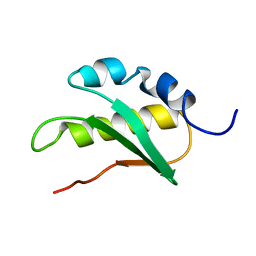 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU-like protein 1, chloroplast | | Authors: | Saio, T, Ogura, K, Kumeta, H, Yokochi, M, Katoh, S, Katoh, E, Inagaki, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The cooperative role of OsCnfU-1A domain I and domain II in the iron sulphur cluster transfer process as revealed by NMR
J.Biochem.(Tokyo), 142, 2007
|
|
8KG3
 
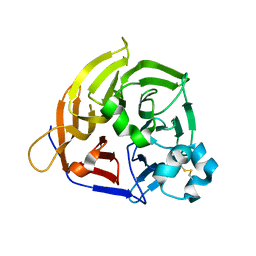 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
2CVO
 
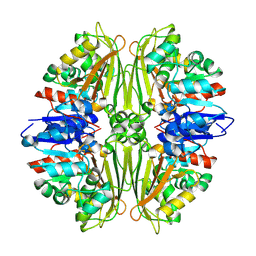 | | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) | | Descriptor: | putative Semialdehyde dehydrogenase | | Authors: | Nonaka, T, Kita, A, Miura-Ohnuma, J, Katoh, E, Inagaki, N, Yamazaki, T, Miki, K. | | Deposit date: | 2005-06-10 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa)
Proteins, 61, 2005
|
|
1WMJ
 
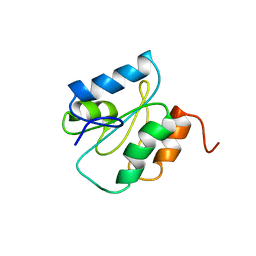 | |
3WRV
 
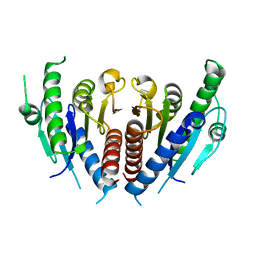 | |
1IRZ
 
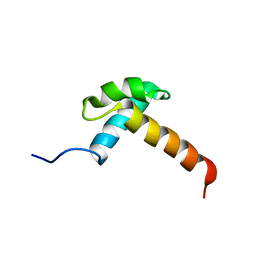 | |
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
3VKW
 
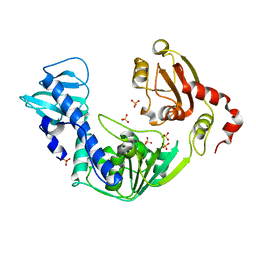 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
3WRY
 
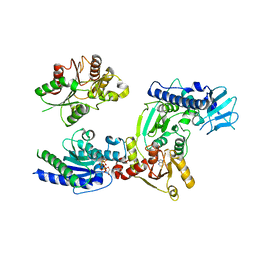 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRX
 
 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
